Chris Johnson to Co-Chair and Host SIAM Computational Science and Engineering Conference
 The SIAM Computational Science and Engineering 2015 Conference will be held March 14-18 at the The Calvin L. Rampton Salt Palace Convention Center, 100 S West Temple, Salt Lake City, UT 84101
The SIAM Computational Science and Engineering 2015 Conference will be held March 14-18 at the The Calvin L. Rampton Salt Palace Convention Center, 100 S West Temple, Salt Lake City, UT 84101We are expecting more than 1500 attendees. A large number of presentations will be given. See the Conference Program for details.
Both Sides of The Brain
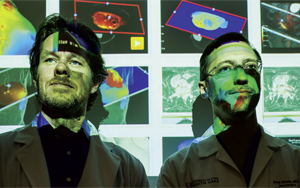 HOW UNIVERSITY OF UTAH HEALTH CARE REMAINS AMONG THE NATION'S BEST IN DEEP BRAIN STIMULATION (DBS)
HOW UNIVERSITY OF UTAH HEALTH CARE REMAINS AMONG THE NATION'S BEST IN DEEP BRAIN STIMULATION (DBS)From University of Utah Clinical Neurosciences Center Convergence (See page 12)
By Jennifer Dobner
It sounds like something straight from a scene in a science fiction film: Surgery that places a set of wires under the skull so that electrical signals can be transmitted to different areas of the brain. It's called DBS, or deep brain stimulation. And if the idea of it seems a bit wince-inducing or scary, then understanding the power of what it can do - quiet the tremors associated with Parkinson's disease and other brain disorders - will likely wash away any patient's fears.
NVIDIA Center of Excellence Renewed
 The NVIDIA Corporation, the worldwide leader in visual computing technologies has renewed the University of Utah's recognition as a CUDA Center of Excellence, a milestone that marks the continuing of a significant partnership, starting in 2008, between the two organizations.
The NVIDIA Corporation, the worldwide leader in visual computing technologies has renewed the University of Utah's recognition as a CUDA Center of Excellence, a milestone that marks the continuing of a significant partnership, starting in 2008, between the two organizations.NVIDIA® CUDA™ technology is an award-winning C-compiler and software development kit (SDK) for developing computing applications on GPUs. Its inclusion in the University of Utah's curriculum is a clear indicator of the ground-swell that parallel computing using a many-core architecture is having on the high-performance computing industry. One of twenty-two centers, the University of Utah was the second school to be recognized as a CUDA Center of Excellence along with the University of Illinois at Urbana-Champaign. Over 50 other schools and universities now include CUDA technology as part of their Computer Science curriculum or in their research.
U of U team finishes study of massive, mysterious explosion
 (KUTV) A team of researchers from the University of Utah is wrapping up an exhaustive five-year study looking into a mysterious explosion of a semi-truck in Spanish Fork Canyon back in 2005.
(KUTV) A team of researchers from the University of Utah is wrapping up an exhaustive five-year study looking into a mysterious explosion of a semi-truck in Spanish Fork Canyon back in 2005.The truck was packed with 35,000 pounds of mining explosives. It blew up after the truck rolled over, leaving a massive crater, 70-feet wide and 30-feet deep.
There were no fatalities but explosions like the one that happened on Aug. 10, 2005 are extremely rare. But the team of researchers is determined to prevent this type of incident from happening again.
Martin Berzins appointed Member of ASCAC
 Martin Berzins has been appointed a member of the Advanced Scientific Computing Advisory Committee (ASCAC). The committee provides advice and recommendations on scientific, technical, and programmatic issues relating to the ASCAC Program.
Martin Berzins has been appointed a member of the Advanced Scientific Computing Advisory Committee (ASCAC). The committee provides advice and recommendations on scientific, technical, and programmatic issues relating to the ASCAC Program.The Advanced Scientific Computing Advisory Committee (ASCAC), established on August 12, 1999, provides valuable, independent advice to the Department of Energy on a variety of complex scientific and technical issues related to its Advanced Scientific Computing Research program.
Learn more about ASCAC
Simulations Aimed at Safer Transport of Explosives
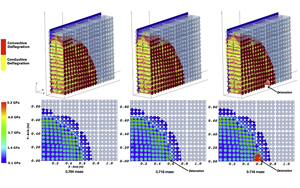 Credit: Jim Collins - Argonne Leadership Computing Facility
Credit: Jim Collins - Argonne Leadership Computing FacilitySee original article: "Simulations Aimed at Safer Transport of Explosives"
In 2005, a semi-truck hauling 35,000 pounds of explosives through the Spanish Fork Canyon in Utah crashed and caught fire, causing a dramatic explosion that left a 30-by-70-foot crater in the highway.
Fortunately, there were no fatalities. With about three minutes between the crash and the explosion, the driver and other motorists had time to flee. Some injuries did occur, however, as the explosion sent debris flying in all directions and produced a shock wave that blew out nearby car windows.
UDCC Open House
 Wednesday, September 17th 2014
Wednesday, September 17th 201410 am to 3 pm
University of Utah
Warnock Engineering Building, Catmull Gallary
72 So. Central Campus Dr.
The first UDCC open house will bring together our consortium partners and engineering students to a single venue. Partners interested in sponsoring student internships through the new Data Center Engineering Certificate will be present for questions, and students will have the opportunity to hear from and engage with some of our nation's leading experts in the field. You can visit our website or email us for more information.
2014 Summer Course on Image-based Biomedical Modeling (IBBM)
The Image-Based Biomedical Modeling (IBBM) summer course was held from July 14 to July 24 in the Newpark Hotel, Park City, Utah.
The two-week summer course hosted 39 participants this year: 31 graduate students, 1 MD/PhD student, 2 postdoctoral fellows, 3 junior faculty, and 2 developers from a research laboratory / industry. Participants came from 24 institutions, including 4 from universities in Belgium and England. After the first week of common classes, participants were divided into two tracks: Bioelectricity (10 participants) and biomechanics (29 participants).
 IBBM is a dedicated two-week course in the area of image-based modeling and simulation applied to bioelectricity and biomechanics, providing participants with training in the numerical methods, image analysis, visualization, and computational tools necessary to carry out end-to-end, image-based, subject-specific simulations in either bioelectricity or orthopedic biomechanics. The course focuses on using freely available, open-source software developed under the research of the CIBC (P41 GM103545) and FEBio suite (RO1 GM083925). Students use this software to learn and apply the complete dataflow pipeline to particular sets of data with specific goals.
IBBM is a dedicated two-week course in the area of image-based modeling and simulation applied to bioelectricity and biomechanics, providing participants with training in the numerical methods, image analysis, visualization, and computational tools necessary to carry out end-to-end, image-based, subject-specific simulations in either bioelectricity or orthopedic biomechanics. The course focuses on using freely available, open-source software developed under the research of the CIBC (P41 GM103545) and FEBio suite (RO1 GM083925). Students use this software to learn and apply the complete dataflow pipeline to particular sets of data with specific goals.
Understanding the Morphology of Brain Disorders
Advances in medical imaging devices, such as magnetic resonance imaging (MRI), have led to our ability to acquire detailed information about the living human brain, including its anatomical structure, function, and connectivity. However, making sense of this complex data is a difficult task, especially in large imaging studies that may include hundreds or even thousands of participants. This is where computer science can play an important role. Image analysis algorithms can automatically quantify properties of the brain, such as the size of brain structures, or the functional activity in different brain regions. This provides neuroscience researchers with insights into how the brain functions and what abnormalities are present in diseased brains.
Neuro Stimulation
 |
| Transcranial Magnetic Stimulation (TMS) of the human motor cortex. |
