Software Dissemination as a Form of Science and Technology Dissemination
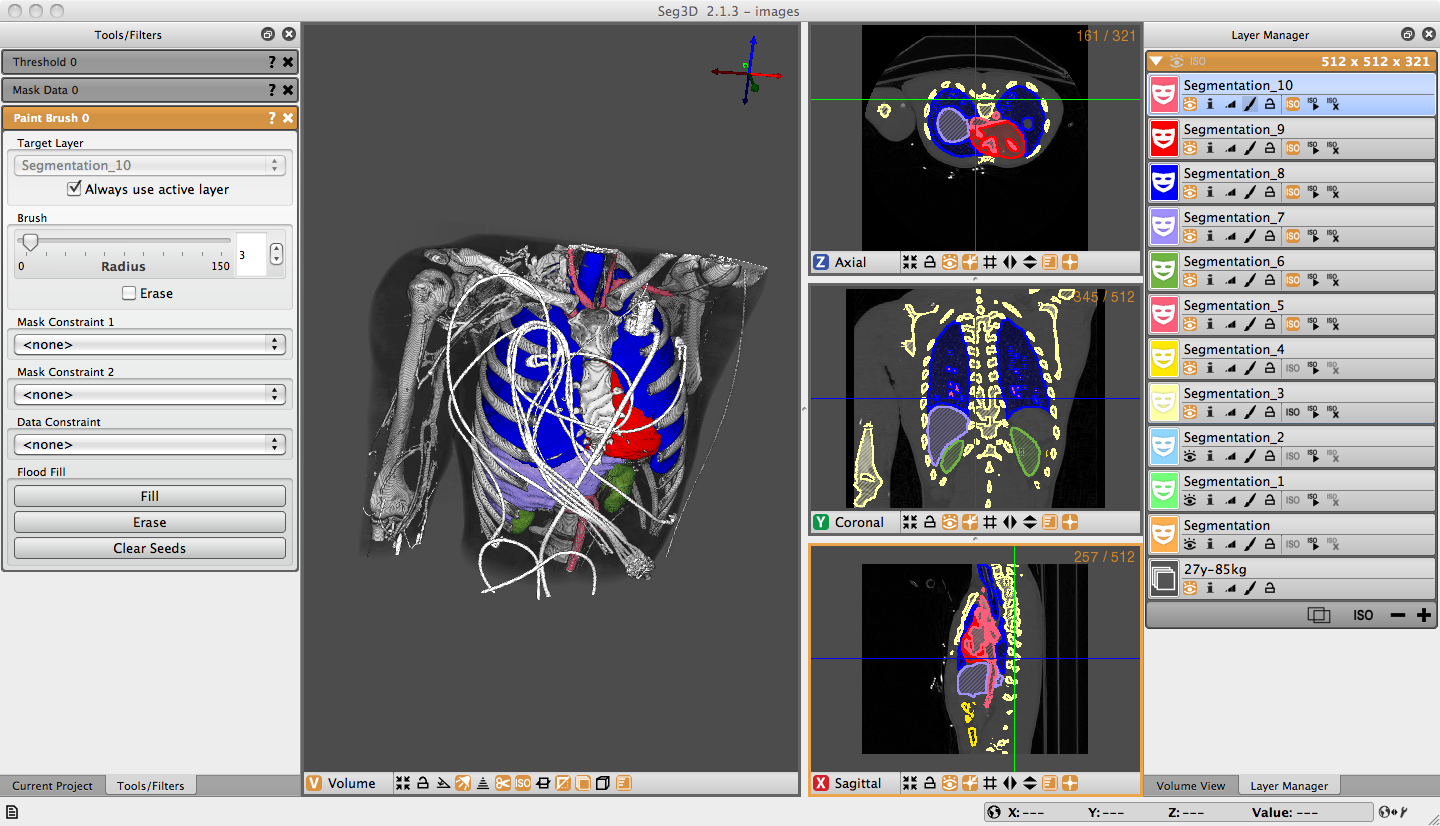
The origins of our success in developing widely used software tools lie in a set of strategies for algorithm research and software development. One such strategy is the production of software tools with low barriers to entry. This entails the release of documented, tested, complete applications that do not require learning new programming languages or complex, architecture-specific build environments. We also continue to follow an initiative to create a suite of lightweight, stand-alone applications, directed at specific tasks of common interest across a wide set of disciplines. The result is a set of programs, such as Seg3D, with large and growing user bases.
Implementing this strategy with the limited resources of a P41 Center has required the development of algorithms that are robust, general purpose, and designed to operate in a wide variety of scenarios, while remaining openly customizable for particular fields or settings. The same need for efficiency drives another important aspect of our strategy: the development of an integrated software infrastructure that allows the same basic algorithms and associated software modules to be used together as part of a user-defined workflow (SCIRun) or separately and in combination with software from other sources (Seg3D, ImageVis3D, BioMesh and map3d). We have yet to achieve a completely interchangeable architecture for this versatile integration; this is a major goal of the next funding period.
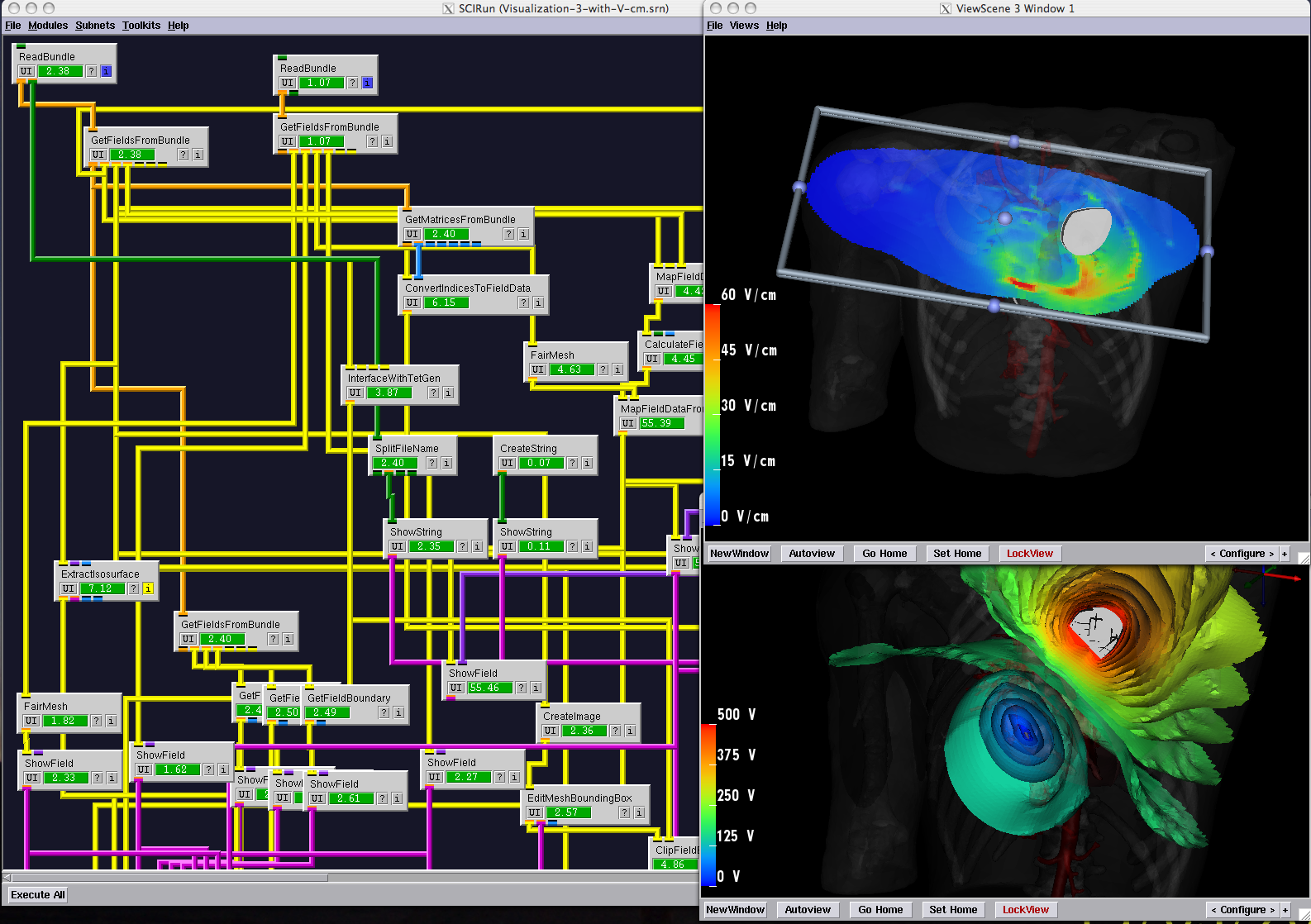
The NIH Center for Bioelectric Field Modeling, Simulation, and Visualization officially began on September 15, 1999. During the first five years, the Center focused on creating an extensible, scalable, scientific problem-solving environment (PSE) and on developing corresponding research to solve real-world problems related to bioelectric fields. To accomplish this goal, we conducted research and development in advanced modeling, simulation, and visualization methods for solving bioelectric field problems; we also created BioPSE, an extension to the existing SCIRun, which is a modular, extensible, integrated software problem-solving environment for bioelectric field problems. The SCIRun software supported interaction among the modeling, computation, and visualization phases of bioelectric field simulation, with BioPSE providing specific extensions for the Center.
The structure of SCIRun resembles the dataflow structure originally developed in a variety of visualization applications (e.g., AVS, Khoros, and Data Explorer), and at the time, its development represented a new approach to addressing the much broader range of computation needs in biomedical research. We imagined SCIRun as a "computational workbench" in which a user selects tools (software modules) from a set of drawers (categories) and connects them to create a workflow (a network) that performs all the necessary steps.
The problem of mesh generation has been widely studied, as a hybrid field of interest to the scientific, engineering, and computer science communities. In each of these fields, meshes are used to compute numerical approximations to solutions of partial differential equations. To do so, continuous mathematics are replaced with a discrete analogue, most commonly to facilitate the finite element method (FEM).
Tens of thousands of users have downloaded the SCIRun and BioPSE software and used it in their bioelectric field research, including cardiac electro-mechanical simulation, ECG/EEG forward and inverse calculations, deep brain stimulation, electromyography calculation, and the determination of the electrical conductivity of anisotropic heart tissue. Users have also employed SCIRun for the visualization of breast tumor brachytherapy, computer-aided surgery, teaching, and a number of non-biomedical applications.
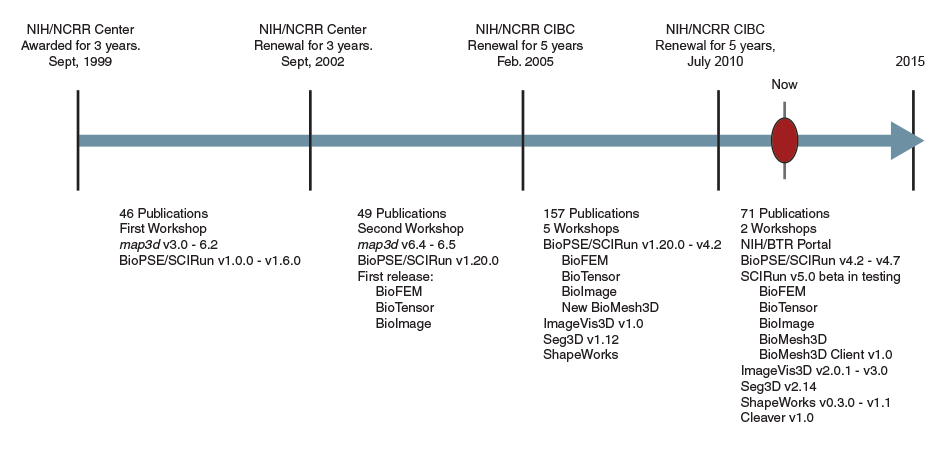 |
| CIBC software dissemination timeline |
Creating and disseminating this integrated, efficient, modular, extensible problem-solving environment for bioelectric field problems was a tremendous challenge, but it is one that we successfully met in 2003(see timeline). After successfully releasing SCIRun and BioPSE, we began to learn from researchers how they were using these tools and what additional capabilities they required. Feedback indicated that the dataflow system of modules (connected by data pipes) was often too complicated for the average biomedical research laboratory, which was accustomed to simple pull-down-menu interfaces seen in typical desktop systems. At our 2003 External Advisory Board meeting, we proposed new approaches to reduce the complexity of the SCIRun interface in order to expand our user base and impact. By 2004 we had created easy-to-use, application-specific software programs based upon SCIRun. We called these programs "PowerApps."
PowerApps looked like dedicated applications but were essentially wrappers around a specifically designed dataflow network. The user saw only the parameters relevant to the task and in a context that could be as domain-specific as necessary. Users reported that their productivity improved because of the lack of distractions and the natural mapping of workflow to user interface.
However, the workflow was driven by the underlying dataflow, a process architecture that provides many advantages but also presents some challenges. In order to overcome these challenges, in 2005 the Center redesigned the package, moving away from a strict link to dataflow and developing in its place truly independent, standalone applications. As part of this move to more independent applications, the Center also re-engineered SCIRun to separate the underlying filters or modules from the dataflow interface. In this way, the basic algorithms and data structures in SCIRun remained accessible in libraries that could then form the basis of new applications, independent of dataflow.
During this time, the Center also integrated third-party packages into its tools. One example is the Insight Toolkit (ITK), a public-domain library of image processing tools created by a consortium and funded by the National Library of Medicine and NIH. Professor Whitaker is a founding member of the Insight Consortium and active in ITK development through his participation in the NAMIC and in the National Center for Biomedical Computing (PI‚R. Kikinis, Harvard University).
StandAlone Applications: Seg3D as an Example
Seg3D is currently used by a large community of researchers. Its applications include the assessment of pulmonary fibrosis (anatomical structure), tracking lesions for macular degeneration, characterization of mice osteochondroma, segmentation in pediatric cardiology in cases of Marfan and Loeys-Dietz Syndromes, and outlining femoacetabular impingement.
Secondary Dissemination by Collaborators
The combination of the modular build of Seg3D and the use of the MIT license has allowed several collaborators to integrate Seg3D into their software applications along with modifications to Seg3D. Examples of this secondary use are the Center for Arrhythmia Research and MAnagement (CARMA), which has created the Coreview system, a software tool developed using Seg3D at MRI image segmentation in support of image-guided ablation for cardiac afibrillation. Likewise, Numira Biosciences, a private CRO, specializes in small animal imaging in support of research and drug development. The software used at Numira is based on the Seg3D package and is used on hundreds of microCT volumes every month.In February 2012, the Seg3D software tool was demonstrated at the SPIE Medical Imaging Conference - Live Demonstration Workshop, and was used to segment MRI cardiac images at the International Symposium on Biomedical Imaging (ISBI) - Challenge Workshop in May 2012. Every year, CIBC staff organizes two hands-on, one-day tutorials as part of its international conferences to teach scientists how to use our software package with their own data.
Benefits to Other Local Software Efforts: Fluorender
The CIBC infrastructure benefits not only from our dissemination efforts, but also from the software created by other research projects. An example of this is the Fluorender software. FluoRender is an interactive rendering tool for confocal microscopy data visualization, created under funding from R01 MH092256-01 and R01 GM098151-01. It combines the renderings of multi-channel volume data and polygon mesh data, where the properties of each dataset can be adjusted independently and quickly. The tool is designed especially for neurobiologists, and it helps them better visualize the fluorescent-stained confocal samples.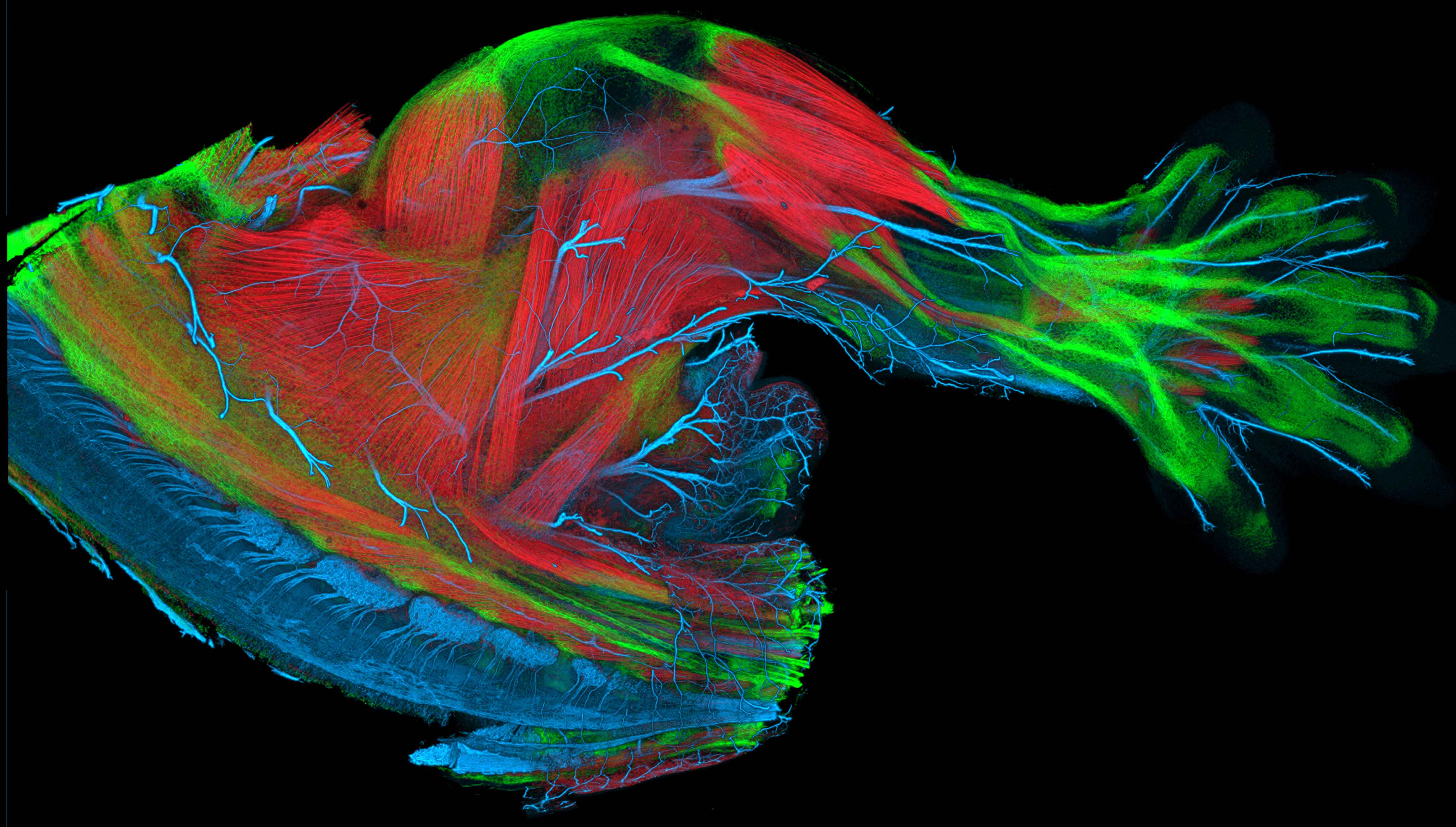 |
| FluoRender is an interactive rendering tool for confocal microscopy data visualization. It combines the renderings of multi-channel volume data and polygon mesh data, where the properties of each dataset can be adjusted independently and quickly. The tool is designed especially for neurobiologists, and it helps them better visualize the fluorescent-stained confocal samples. |