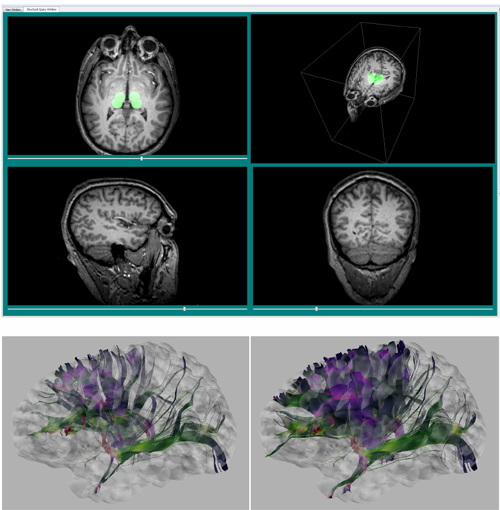
In collaboration with Dr. Don Tucker and his colleagues at Electrical Geodesics Inc (EGI) and the University of Oregon, this DBP is concerned with improving our ability to reconstruct and visualize neuroelectric sources (source localization) from EEG measurements and also our ability to stimulate specific brain regions using electrodes attached on to the scalp of the subject (transcranial direct current stimulation, tDCS).
In collaboration with Dr. Don Tucker and his colleagues at Electrical Geodesics Inc (EGI) and the University of Oregon, this DBP is concerned with improving our ability to reconstruct and visualize neuroelectric sources (source localization) from EEG measurements and also our ability to stimulate specific brain regions using electrodes attached on to the scalp of the subject (transcranial direct current stimulation, tDCS).For both research and clinical practice, EEG is a cost-effective tool to understand and excite brain activity. EEG advances have significantly improved the spatial resolution of source estimates and offer the promise of precise spatio-temporal monitoring and stimulation of cortical brain activity. By itself, high-resolution EEG would be affordable even for small hospitals in remote locations and could be easily managed by technicians in the field.
Within the broad scope of research activity in the field of neural source imaging and stimulation, Dr. Tucker and CIBC have chosen a selected subset of potential projects with the goal of maximizing the potential impact of our collaboration in terms of: 1) improving near-term translational uses of EEG-based source ocalization; 2) moving closer to determining the true resolution possible with EEG; 3) improving the capabilities of our other neuroscience collaborators; and 4) being able to stimulate specific brain regions in a predefined manner and testing these results to address specific clinical problems.
The algorithms and tools developed in this project will be of immediate use to EGI and Dr. Tucker, and we expect to be able to achieve immediate and broad-based dissemination through their base of more than 400 users of their technology as well as CIBC's software dissemination mechanisms. In addition we believe that our ability to collaboratively begin to assert the true potential resolution of EEG in source imaging and noninvasive brain stimulation could make a very positive contribution to the adoption of these relatively low-cost technologies in many research and clinical laboratories.
Creation and Quantitative Comparison of Brain Connectivity Networks
Given the wide array of imaging tools available for the study of brain connectivity such as diffusion MRI, functional MRI, and EEG, networks are becoming a popular tool to integrate and analyze these multiple datasets. In this vain, we are developing tools that allows reconstruction of brain connectivity networks. The design of our tools also allow the quantitative comparison of multiple networks.Structural brain connectivity networks can play an important role in solving the EEG source localization problem. These networks or 'connectome matrices' are useful as priors for stabilizing the source localization inverse solution. Therefore, the accuracy of the reconstruction of connectivity networks directly impacts the quality of the corresponding inverse solution. In this work, we describe brain networks as graphs, where the nodes correspond to anatomical regions, and the edges correspond to anatomical or functional connectivity between regions. Additionally, we developed a metric that allows the quantitative comparison of networks from a single patient. This metric allowed us to examine the impact of imagining variables and algorithmic parameters on network accuracy.
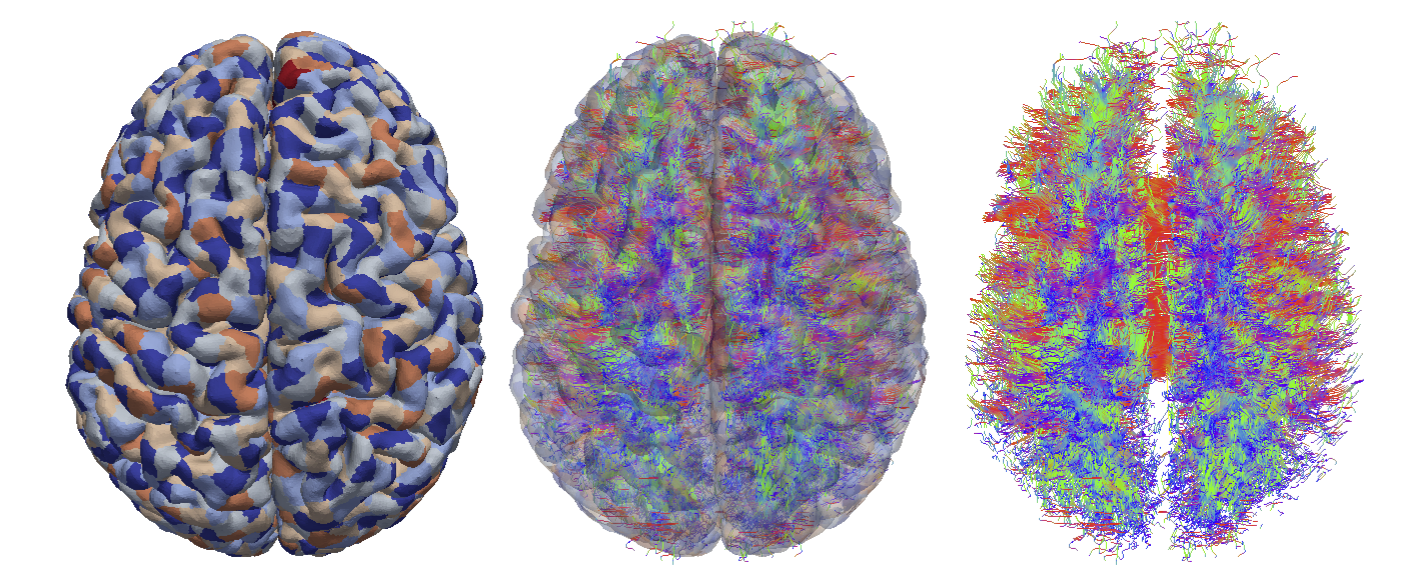 |
| From left to right: Cortical surface mesh showing brain parcellation, tract streamlines, and tract streamlines and cortex, superimposed. |
Creating the Brian Connectivity Network: This study was performed on in vivo image data collected from a healthy subject by Dr. Tucker's group at the Lewis Center of Neuroimaging at the University of Oregon. The first part of this work was concerned with white matter tract reconstruction. First, we fit an orientation distribution function (ODF) to the data in every brain voxel. We subsequently extracted white matter fiber orientations using the tensor decomposition approach presented by Jiao et al1. The fiber orientations were integrated using a deterministic fiber tractography algorithm to yield white matter tracts for the entire brain.
Once white matter tracts were computed, we proceeded with reconstruction of brain connectivity graphs, dividing the brain cortex into surface patches that define the graph vertices. The weighted edges were determined by counting the number of tract streamlines that connect any two cortical surface patches.
Comparing Brain Networks: In the second part of this research, we have addressed the fundamental problem of the comparison of two brain connectivity networks. We created a novel metric, called the 'graph diffusion distance', for comparing two brain networks having the same number of nodes with known correspondences. Our approach is based on measuring the average similarity of diffusion processes specified by the two adjacent matrices. We compute the matrix exponential, or heat diffusion kernel, for the graph Laplacian matrices for each brain network, which requires repeatedly solving the heat diffusion problem with initial conditions localized to single vertices. We define the graph diffusion distance as the Frobenius norm of the difference of the two heat diffusion kernels, at the diffusion time yielding the maximum difference. This metric captures information about the relative effects of edges on the overall diffusion across the graph. In this case we used it to study the effect of q-space undersampling on structural connectivity.
A Tool for the Visual Exploration Multimodal Data and Large Tractographies
After developing a tool capable of fusing these data in a single, coherent way, an additional visualization component was added to adapt the existing multimodal data visualization tool and allow the interaction with larger datasets. This new suite of tools is capable of visualizing arbitrarily large white matter fiber tractography datasets through a streaming renderer. Additional improvements have been made to prune these data by limiting the overall length of tracts shown. User-driven filtering has also been implemented: the figure below shows how users may select regions of the cortex to highlight fiber bundles innervating those locations. Similarly, it demonstrates how interaction with structural MRI data is used to highlight white matter tracts passing through user-defined regions in this separate imaging modality.
Visualization and Analysis of Brain Anatomical Connectivity for Improved EEG Source Localization
The focus of this project was to allow groups such as the Tucker Laboratory to visually explore the multimodal datasets resulting from the integration of EEG signals with the various structural and connectivity datasets. By allowing users to interactively explore data consisting of EEG, MRI, white-matter tractography, and their derived data products, we have enabled scientists to more completely understand how brain structures may influence source localization. In particular, by using structural information gleaned from multimodal data to create and manipulate the connectivity networks, this tool enables the exploration of the effects of onnectivity on the source localization problem.The CIBC contributions to date have included the following:
- We demonstrate that the ability to query collections of timefrequency planes with user-generated examples provides scientists with an effective new tool for the exploration of EEG data,
- We present a novel technique for the exploration of timefrequency patterns through the use of filtered, normalized cross-correlogram images,
- We show that our system enhances the analysis of EEG data during working memory studies, expediting the validation of stated hypotheses.
transcranial Direct Current Stimulation (tDCS)
Transcranial current stimulation (tDCS) is a technique that aims to stimulate the brain by injecting low amplitude currents through the electrodes on the scalp. tDCS has seen growing attention during the last decade both as a therapy for brain disorders and as a stimulus to increase motor function. For clinical purposes such as supporting treatment of mood disorders or enhancing learning, it is desirable to deliver the lowest amplitude electrical currents possible to a particular brain region in a specific direction without widespread brain activation. Thus, a precise understanding of energy deposition is required.Simulation of Transcranial Direct Current Stimulation for Realistic Human Head Models
The goal of our first project was to develop and validate a computational pipeline capable of optimizing electrode configurations and current injection patterns to stimulate only specific desired brain regions. In tDCS simulations, solving of the tDCS forward problem requires a geometric model of the head, typically from medical image data, as a volume conductor for realistic modeling of electrical field propagation. In general, this electrical field propagation (volume conduction) depends mainly on tissue conductivities that are only roughly known in vivo for many of the involved tissues e.g. skull, skin, grey and white matter of the brain, cerebrospinal fluid (CSF), etc. We present a pipeline to compute tDCS simulations for realistic volume conductor models using a combination of open source software tools that include ImageVis3D, Seg3D, BioMesh3D, and SCIRun, all products of CIBC. Our proposed pipeline is capable of generating realistic head models for finite element model (FEM) simulations using an adaptive meshing approach (BioMesh3D). The implemented SCIRun modules were able to model arbitrary shapes of electrodes and configurations as well as realistic boundary conditions specified by the complete electrode model. For exhaustive tDCS simulations, e.g. for the optimization of electrode configurations and injection current patterns, we used a technique called the transfer matrix approach to reduce the computational effort.Patch Electrodes Versus High Density Electrode Arrays
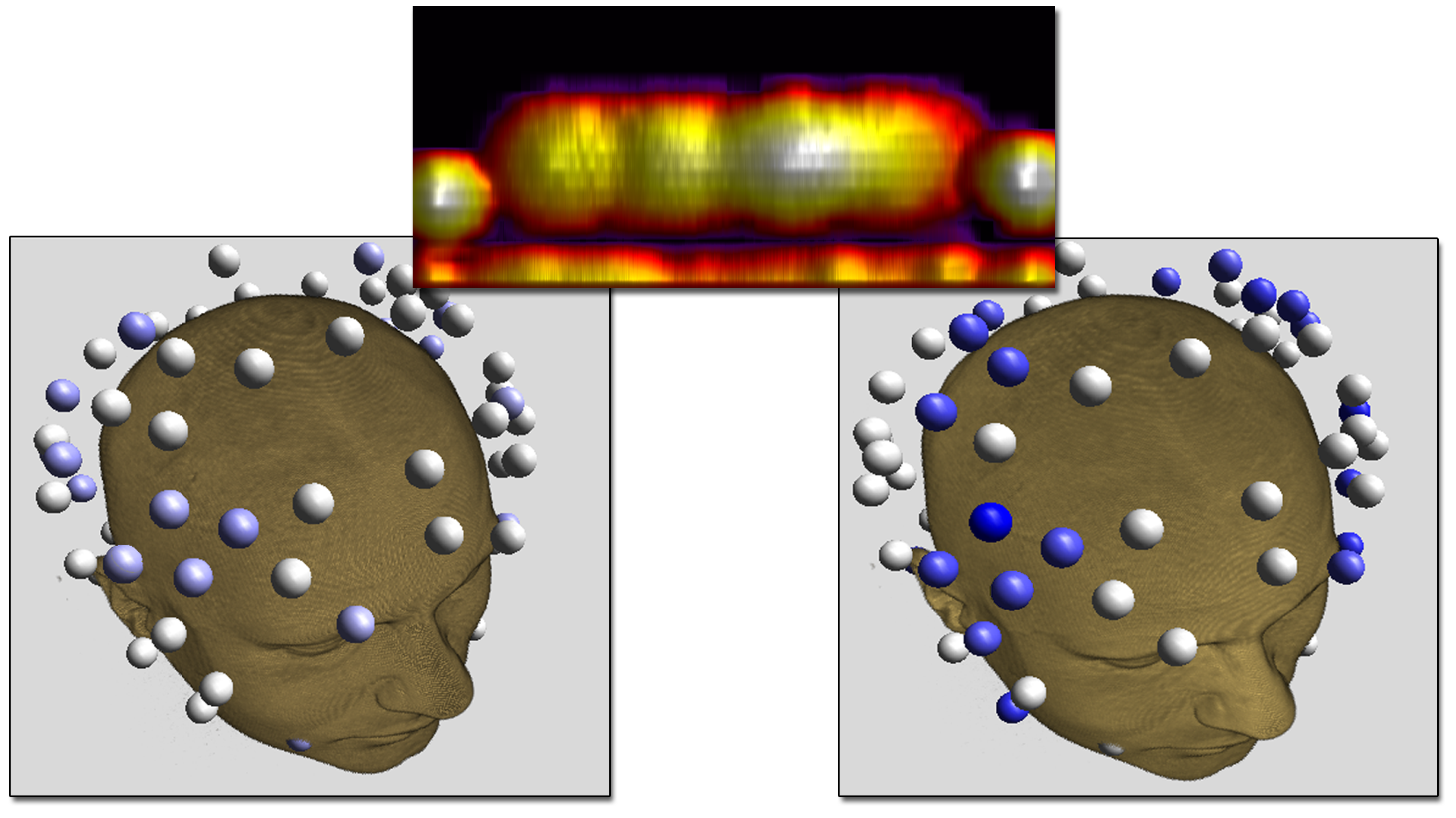
Electrode shape, electrode montage, and electric stimulation parameters all affect how cortical brain areas are stimulated. Therefore, the settings of tDCS stimulation are crucial for both accuracy and the effect of the stimulation as well as the safety of the subject. Typically large patch electrodes are used to stimulate coarsely targeted brain regions while minimizing electrical currents injected on the scalp. However, the utility of stimulating more focal brain regions while still guaranteeing safety is of medical interest. The aim of our studies was to compare performance of patch electrodes to that of dense electrode arrays in terms of activated cortex and safety, using the realistic head model described earlier.Using high-density (HD) electrode arrays increases our ability to control the current patterns in the brain. We were able to confirm that HD electrode arrays offer more flexible ways to stimulate particular brain regions compared to the commonly used patch electrode.
Our results show that HD electrode arrays are capable of mimicking spatially broad (figure above, left) but also focal brain stimulations (figure above, right) while guaranteeing the subject's safety. However, it remains difficult to manually determine the best electrode injection pattern to maximize the stimulation in that region.
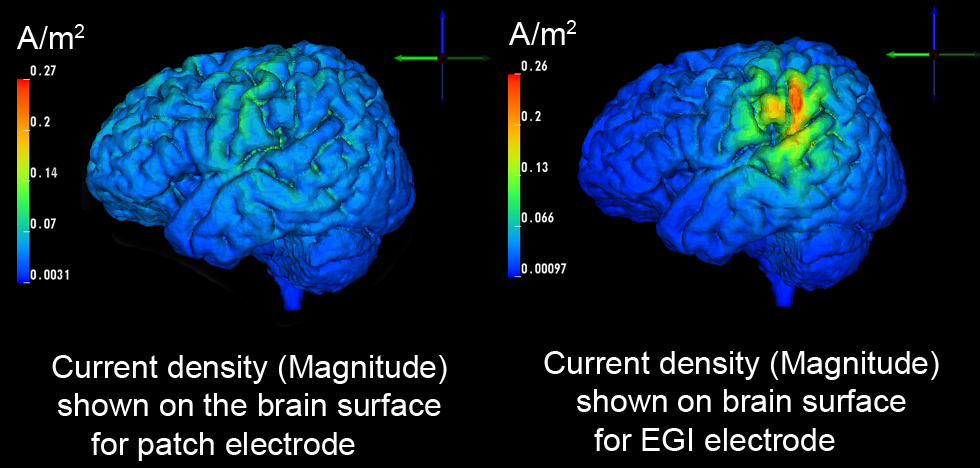 |
| Comparision of stimulation effect using a patch electrode (left, 5x5 cm surface area) vs. HD electrodes (right, 1.2 cm surface area) |
Optimization of Current Stimulus Patterns
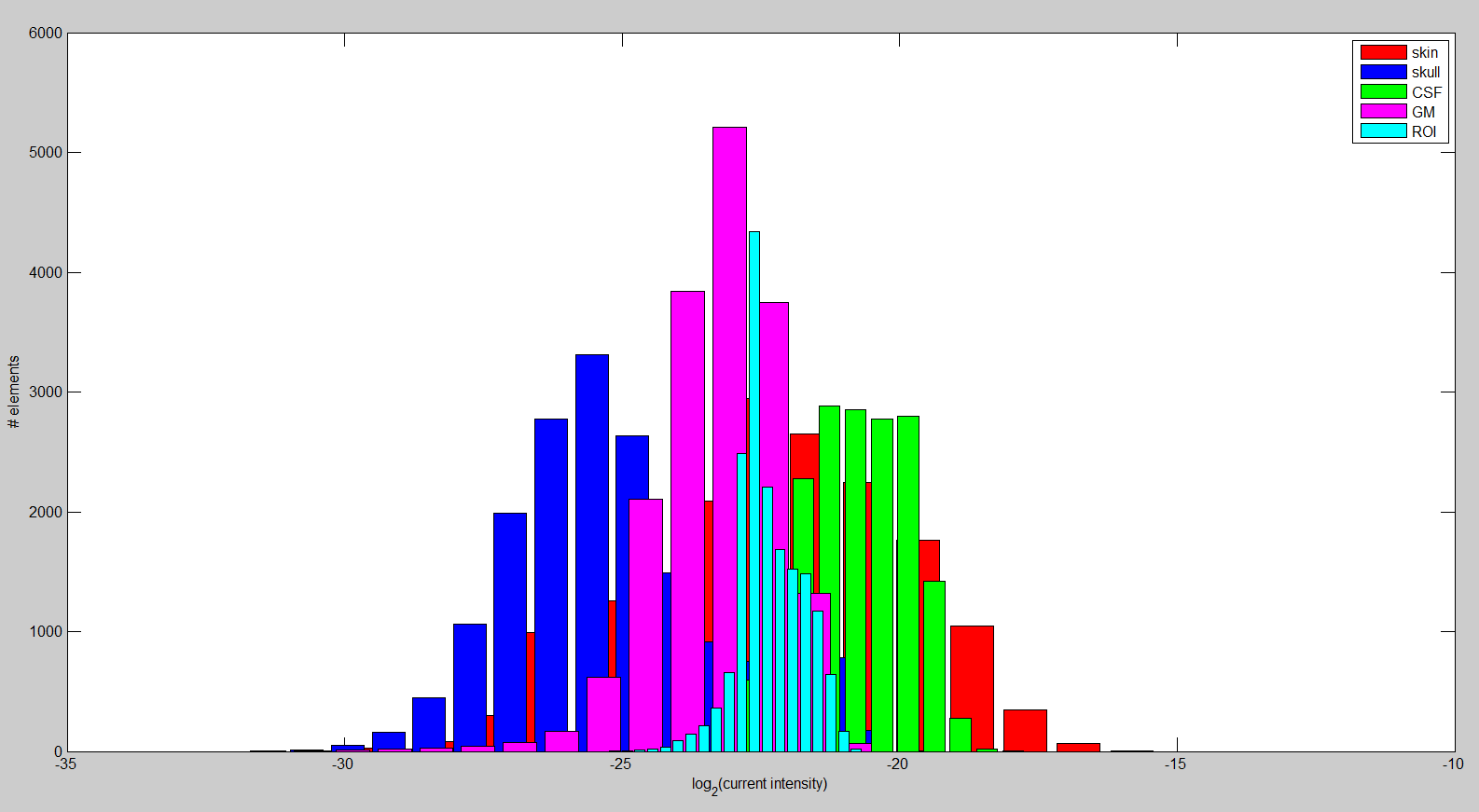
In the last aspect of the research on tDCS, we want to maximize the directional current in the brain region of interest (ROI) by formulating a multiconstraint optimization problem. We imposed constraints on the total injected current, each electrode current, and total power in the brain outside ROI to ensure the safety and comfort of the subject. After discretization of the realistic head model described above, we computed the required matrices to set up the convex optimization problem. The linear relationship between mesh node potentials and electrode current injection allows us to precompute a transfer matrix that is used during optimization. This procedure reduces the computation time dramatically, leading to the possibility of interactively changing ROI or the desired stimulation direction.Overall, the results suggest that we are capable of stimulating the ROI for an arbitrary direction: The left hand panel of figure above depicts the currents that are running through the ROI. Additionally, it is important to show that the solution appears to be focal. In the right hand panel of the figure, the histogram of the current intensity in different regions is shown. It appears to be higher and rather focal compared to most other tissues. Furthermore, the current intensity in the ROI is larger on average compared to the rest of the brain. The difference increases considerably when we choose a ROI close to the head surface. Lastly, we are investigating the effect of the anisotropy in very complex volume conductor models2 on the current optimization. Our findings suggest that skull and white matter anisotropy change the solution significantly depending on the ROI and desired direction selection.
All the algorithms and tools developed in a collaborative effort with our EGI colleagues are of immediate use to EGI and Dr. Tucker's group. We expect to be able to achieve immediate and broad-based dissemination through their base of more than 400 users of their technology. In addition we believe that our ability to collaboratively begin to assert the true potential resolution of EEG in source imaging and noninvasive brain stimulation could make a very positive contribution to the adoption of this relatively low-cost technology in many research and clinical laboratories.
References
1. F. Jiao, Y. Gur, C.R. Johnson, and S. Joshi. Detection of crossing white matter fibers with high-order tensors and rank-k decompositions. In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI 2011), volume 6801 of Lecture Notes in Computer Science (LNCS), pages 538–549, 2011.2. M. Dannhauer, R.S. MacLeod, S. Guler, D. Tucker, S. Turovets, C. Mattson, and D.H. Brooks. Comparison of patch electrodes and high density electrode arrays regarding volume conduction in tDCS using a realistic head model. In Human Brain Mapping, 2013.