Mathematical Technique Reveals Predictive DNA Patterns That Other Methods Missed
Nearly anyone touched by ovarian cancer will tell you: it's devastating. It's bad enough that cancer in almost 80 percent of patients reaches advanced stages before diagnosis, and that most patients are expected to die within five years. But just as painfully, roughly one quarter of women diagnosed have no warning that they are resistant to platinum-based chemotherapy, the main line of defense, nor that they will likely have 18 months to live.
Now, University of Utah scientists have uncovered patterns of DNA anomalies that predict a woman's outcome significantly better than tumor stage. In addition, these patterns are the first known indicator of how well a woman will respond to platinum therapy. Published in the journal PLOS ONE, the patterns were discovered by using a new mathematical technique in the analysis of DNA profiles from the Cancer Genome Atlas, a national database containing data from hundreds of ovarian cancer patients.
"We believe this is a first step toward bringing ovarian cancer into the age of precision medicine," says team leader Orly Alter, Ph.D., USTAR associate professor of bioengineering, adjunct associate professor of human genetics, and faculty member of the Scientific Computing and Imaging Institute. Pending experimental revalidation in the clinic, the patterns could be the basis of a personalized prognostic and diagnostic laboratory test. This test would predict both the patient's survival and the tumor's sensitivity to platinum-based chemotherapy, and doctors could tailor treatment accordingly.
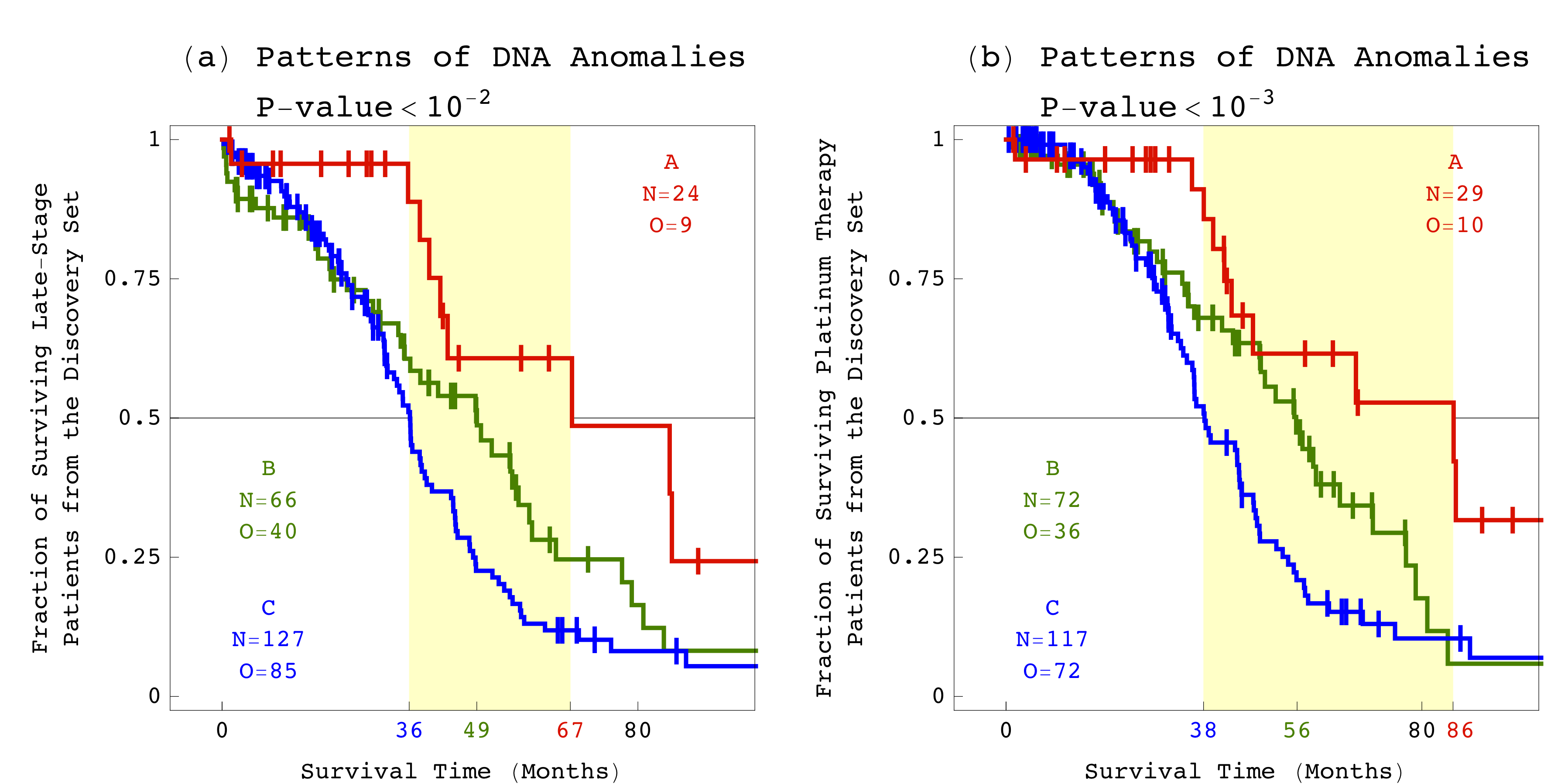
For example, among patients that were diagnosed at late stages, the DNA patterns distinguished short-term survivors, with a median survival time of three years, from long-term survivors, with a median survival time almost twice as long. Among patients treated with platinum-based chemotherapy drugs, the DNA patterns distinguished those with platinum-resistant tumors, with a median survival time of three years, from those with platinum-sensitive tumors, with a median survival time of more than seven years. Alter's team computationally validated the results by using data from independent sets of patients.
"If we have a tool that can more accurately predict survival, and distinguish who is who, we can revamp our entire approach to how we treat patients," says Margit-Maria Janát-Amsbury, M.D., Ph.D., research assistant professor in obstetrics and gynecology, director of gynecologic oncology research at the University of Utah School of Medicine, and faculty member of the Huntsman Cancer Institute. She is collaborating with Alter to bring her team's results to the clinic. "For those with a poor prognosis, we can suggest other therapies, or we can focus on taking measures to improve quality of life."
"What made our discovery possible is our new technique for mathematical modeling," said Alter. "It may very well be that the data needed to better treat cancer are already published. The ovarian cancer data, for example, were published back in 2011. The bottleneck to discovery is in the analysis of the data."
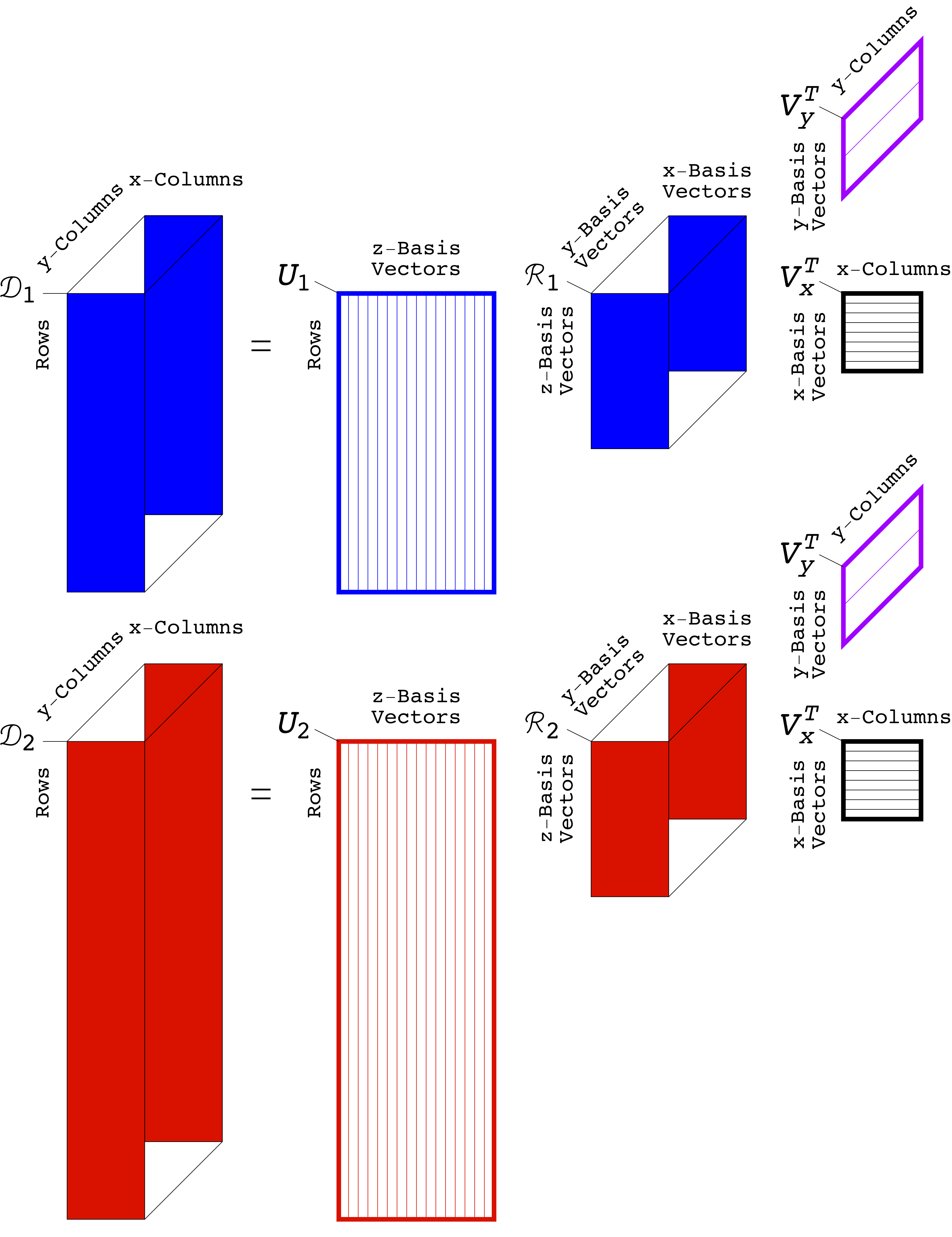
In Alter's Genomic Signal Processing Lab, Ph.D. graduate students and study co-authors Katherine Aiello and Theodore Schomay of the department of bioengineering develop algorithms to uncover patterns in datasets arranged in multidimensional tables, known as tensors. Rather than simplifying the big data, as is commonly done, the algorithms make use of the complexity of the data in order to tease out the patterns within them. Here, for example, by modeling DNA profiles of tumor and normal cells from the same set of patients, they were able to separate the patterns of DNA anomalies – which occur only in tumor genomes – from those that occur in the genomes of normal cells in the body, and from variations caused by experimental inconsistencies.
The algorithms extend a mathematical technique called the singular value decomposition, or SVD. The SVD helps us understand data arranged in two-dimensional tables, known as matrices, by breaking the data down into individual components. In physics, for example, the SVD describes the activity of a prism, which splits white light into its component colors. "It seemed natural that generalizations of the SVD could separate the multidimensional data that arise in personalized medicine into mathematical patterns that have biological meaning," explains Alter, who has a Ph.D. in applied physics.
Alter says her algorithms could just as readily be applied to any type of data. She previously used a similar mathematical technique to uncover new prognostic and diagnostic DNA indicators for patients with glioblastoma, the most common brain cancer in adults. The best predictor of glioblastoma survival prior to Alter's discovery was the patient's age at diagnosis.
Some of the specific genes that Alter's team found to be perturbed in glioblastoma, and in ovarian cancer, may be actively involved in promoting or inhibiting cancer development and progression. Future work will explore whether existing drugs that target these genes are effective in treating these diseases.
To read the April 15, 2015 PLOS ONE article go to:
http://dx.plos.org/10.1371/journal.pone.0121396
This work was supported by the Utah Science, Technology, and Research (USTAR) Initiative, National Human Genome Research Institute (NHGRI) R01 Grant HG-004302, and National Science Foundation (NSF) CAREER Award DMS-0847173.
Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival. Sankaranarayanan P, Schomay TE, Aiello KA, Alter O. PLOS ONE 10 (4), article e0121396 (April 15, 2015); doi: 10.1371/journal.pone.0121396
Contacts:
Orly AlterAssociate Professor, Bioengineering
Adjunct Associate Professor, Human Genetics
Faculty, Scientific Computing and Imaging Institute
Julie Kiefer
University of Utah Health Sciences Public Affairs
News coverage for this article:
U News Center
AAAS EurekAlert!
National Academy of Engineering Podcast and Radio Series