The SCI Institute proudly releases SCIRun version 1.2.0. In direct response to feedback from program users and collaborators, this version contains several new features.
New Features of SCIRun v1.2.0
- Dynamic Compilation and Loading
- PETSc SLES support
- Disjoint package builds
- Improved Documentation
Dynamic Compilation and Loading
DCL provides dynamic compilation of algorithms in SCIRun which reduces compile-to-experiment time. Often called
deferred compilation, the templated C++ code in SCIRun now compiles only when you use it.
PETSc SLES Support
The SolveMatrix module now utilizes PETSc methods and preconditioners for solving linear systems. By making use of robust standards like PETSc, we have improved the range of systems that SCIRun can solve and provided a pathway for additional growth in linear algebra support for future releases.
Disjoint Package Builds
Designed with large site maintenance in mind, SCIRun provides methods to do "disjoint package" builds. Given a research lab with many machines and a central repository of binaries, SCIRun can be deployed much like an emacs or LaTeX. Each workstation can share common binaries and libraries while maintaining individual packages locally.
Improved Documentation
This release provides the first SCIRun step-by-step tutorial. Designed for the first time SCIRun user, the tutorial guides the user through the philosophy and methodology behind the SCIRun dataflow networks and modules. The addition of new networks and datasets with the SCIRun installation will further accelerate and enhance learning to use SCIRun. Finally, we continue to expand the documentation for SCIRun and BioPSE, which is available online via our website, locally by any web browser, and in pdf form for printing. See our documentation page for access.
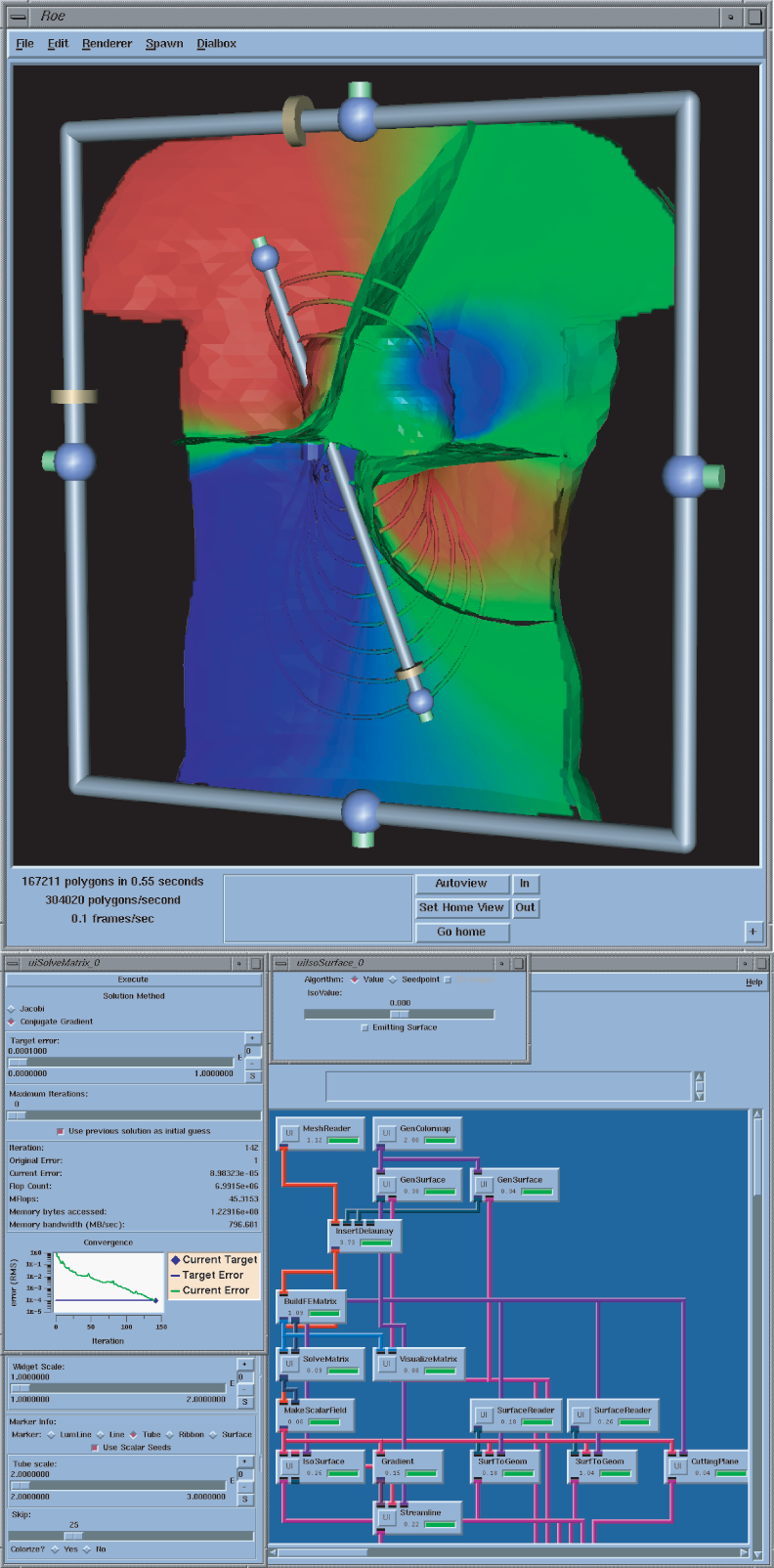 The SCI Institute proudly releases SCIRun version 1.2.0. In direct response to feedback from program users and collaborators, this version contains several new features.
The SCI Institute proudly releases SCIRun version 1.2.0. In direct response to feedback from program users and collaborators, this version contains several new features. 