2012
N.W. Akoum, C.J. McGann, G. Vergara, T. Badger, R. Ranjan, C. Mahnkopf, E.G. Kholmovski, R.S. Macleod, N.F. Marrouche.
“Atrial Fibrosis Quantified Using Late Gadolinium Enhancement MRI is AssociatedWith Sinus Node Dysfunction Requiring Pacemaker Implant,” In Journal of Cardiovascular Electrophysiology, Vol. 23, No. 1, pp. 44--50. 2012.
DOI: 10.1111/j.1540-8167.2011.02140.x
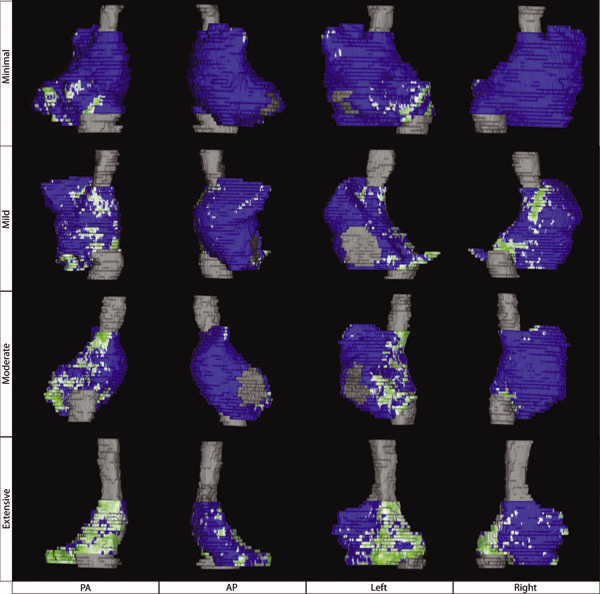
Atrial Fibrosis and Sinus Node Dysfunction. Introduction: Sinus node dysfunction (SND) commonly manifests with atrial arrhythmias alternating with sinus pauses and sinus bradycardia. The underlying process is thought to be because of atrial fibrosis. We assessed the value of atrial fibrosis, quantified using Late Gadolinium Enhanced-MRI (LGE-MRI), in predicting significant SND requiring pacemaker implant.
Methods: Three hundred forty-four patients with atrial fibrillation (AF) presenting for catheter ablation underwent LGE-MRI. Left atrial (LA) fibrosis was quantified in all patients and right atrial (RA) fibrosis in 134 patients. All patients underwent catheter ablation with pulmonary vein isolation with posterior wall and septal debulking. Patients were followed prospectively for 329 ± 245 days. Ambulatory monitoring was instituted every 3 months. Symptomatic pauses and bradycardia were treated with pacemaker implantation per published guidelines.
Results: The average patient age was 65 ± 12 years. The average wall fibrosis was 16.7 ± 11.1% in the LA, and 5.3 ± 6.4% in the RA. RA fibrosis was correlated with LA fibrosis (R2= 0.26; P < 0.01). Patients were divided into 4 stages of LA fibrosis (Utah I: 35%). Twenty-two patients (mean atrial fibrosis, 23.9%) required pacemaker implantation during follow-up. Univariate and multivariate analysis identified LA fibrosis stage (OR, 2.2) as a significant predictor for pacemaker implantation with an area under the curve of 0.704.
Conclusions: In patients with AF presenting for catheter ablation, LGE-MRI quantification of atrial fibrosis demonstrates preferential LA involvement. Significant atrial fibrosis is associated with clinically significant SND requiring pacemaker implantation. (J Cardiovasc Electrophysiol, Vol. 23, pp. 44-50, January 2012)
S.P. Awate, P. Zhu, R.T. Whitaker.
“How Many Templates Does It Take for a Good Segmentation?: Error Analysis in Multiatlas Segmentation as a Function of Database Size,” In Int. Workshop Multimodal Brain Image Analysis (MBIA) at Int. Conf. MICCAI, Lecture Notes in Computer Science (LNCS), Vol. 2, Note: Recieved Best Paper Award, pp. 103--114. 2012.
PubMed ID: 24501720
PubMed Central ID: PMC3910563
J.R. Bronson, J.A. Levine, R.T. Whitaker.
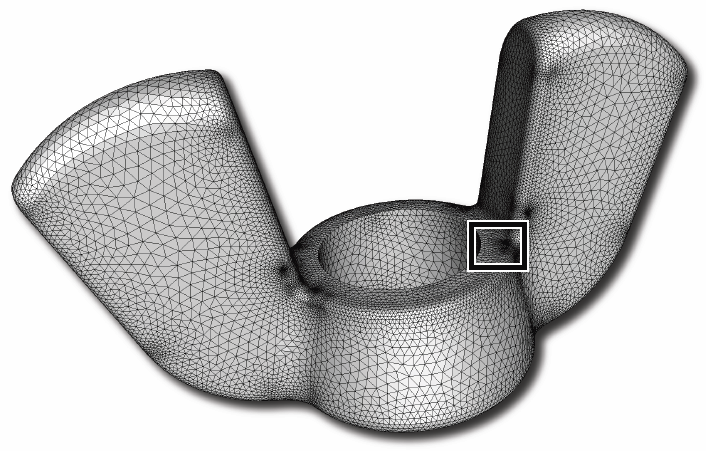
“Lattice Cleaving: Conforming Tetrahedral Meshes of Multimaterial Domains with Bounded Quality,” In Proceedings of the 21st International Meshing Roundtable, pp. 191--209. 2012.

J.R. Bronson, J.A. Levine, R.T. Whitaker.
“Particle Systems for Adaptive, Isotropic Meshing of CAD Models,” In Engineering with Computers, Vol. 28, No. 4, pp. 331--344. 2012.
PubMed ID: 23162181
PubMed Central ID: PMC3499137
C. Butson, G. Tamm, S. Jain, T. Fogal, J. Krüger.
“Evaluation of Interactive Visualization on Mobile Computing Platforms for Selection of Deep Brain Stimulation Parameters,” In IEEE Transactions on Visualization and Computer Graphics, pp. (accepted). 2012.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2012.92
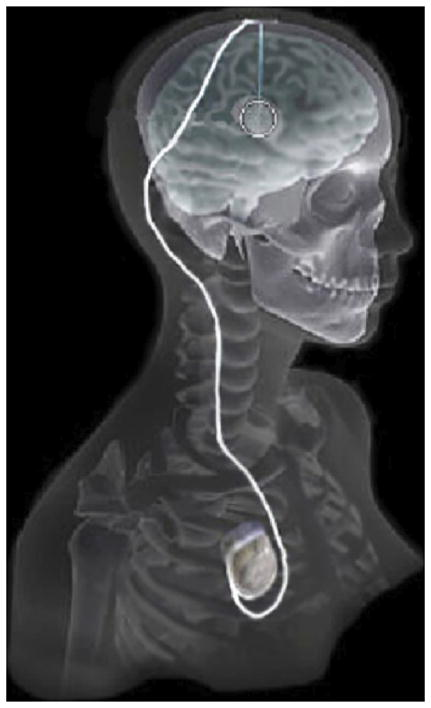
In recent years there has been significant growth in the use of patient-specific models to predict the effects of deep brain stimulation (DBS). However, translating these models from a research environment to the everyday clinical workflow has been a challenge. In this paper, we deploy the interactive visualization system ImageVis3D Mobile in an evaluation environment to visualize models of Parkinson’s disease patients who received DBS therapy. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy; and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independently of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard of care, but were selected in drastically less time. We show how our visualization system can be used to guide clinical decision making for selection of DBS settings.
Keywords: scidac, dbs
A.N.M. Imroz Choudhury, Bei Wang, P. Rosen, V. Pascucci.
“Topological Analysis and Visualization of Cyclical Behavior in Memory Reference Traces,” In Proceedings of the IEEE Pacific Visualization Symposium (PacificVis 2012), pp. 9--16. 2012.
DOI: 10.1109/PacificVis.2012.6183557
Keywords: scidac
M. Dannhauer, D.H. Brooks, D. Tucker, R.S. MacLeod.
“A pipeline for the simulation of transcranial direct current stimulation for realistic human head models using SCIRun/BioMesh3D,” In Proceedings of the 2012 IEEE Int. Conf. Engineering and Biology Society (EMBC), pp. 5486--5489. 2012.
DOI: 10.1109/EMBC.2012.6347236
PubMed ID: 23367171
PubMed Central ID: PMC3651514

M. Datar, P. Muralidharan, A. Kumar, S. Gouttard, J. Piven, G. Gerig, R.T. Whitaker, P.T. Fletcher.
“Mixed-Effects Shape Models for Estimating Longitudinal Changes in Anatomy,” In Spatio-temporal Image Analysis for Longitudinal and Time-Series Image Data, Lecture Notes in Computer Science, Vol. 7570, Springer Berlin / Heidelberg, pp. 76--87. 2012.
ISBN: 978-3-642-33554-9
DOI: 10.1007/978-3-642-33555-6_7
Keywords: Computer Science
B. Erem, P. Stovicek, D.H. Brooks.
“Manifold learning for analysis of low-order nonlinear dynamics in high-dimensional electrocardiographic signals,” In Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 844--847. 2012.
DOI: 10.1109/ISBI.2012.6235680
F.N. Golabchi, D.H. Brooks.
“Axon segmentation in microscopy images - A graphical model based approach,” In Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 756-759. 2012.
DOI: 10.1109/ISBI.2012.6235658
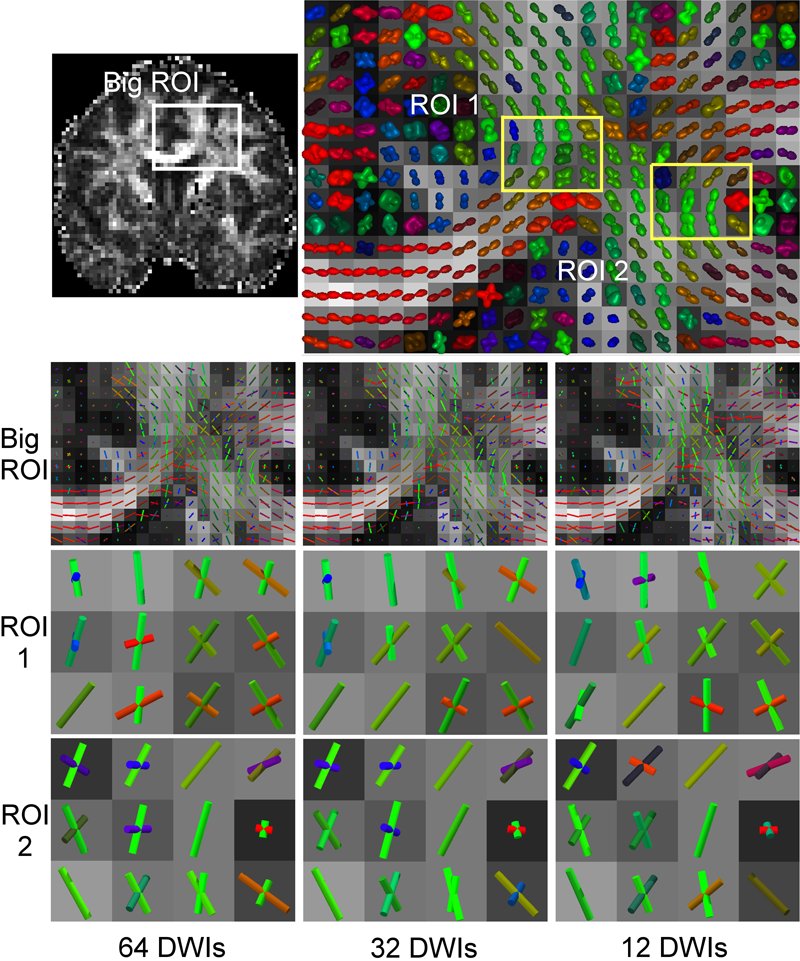
Y. Gur, F. Jiao, S.X. Zhu, C.R. Johnson.
“White matter structure assessment from reduced HARDI data using low-rank polynomial approximations,” In Proceedings of MICCAI 2012 Workshop on Computational Diffusion MRI (CDMRI12), Nice, France, Lecture Notes in Computer Science (LNCS), pp. 186-197. October, 2012.
C.R. Johnson.
“Biomedical Visual Computing: Case Studies and Challenges,” In IEEE Computing in Science and Engineering, Vol. 14, No. 1, pp. 12--21. 2012.
PubMed ID: 22545005
PubMed Central ID: PMC3336198

Computer simulation and visualization are having a substantial impact on biomedicine and other areas of science and engineering. Advanced simulation and data acquisition techniques allow biomedical researchers to investigate increasingly sophisticated biological function and structure. A continuing trend in all computational science and engineering applications is the increasing size of resulting datasets. This trend is also evident in data acquisition, especially in image acquisition in biology and medical image databases.
For example, in a collaboration between neuroscientist Robert Marc and our research team at the University of Utah's Scientific Computing and Imaging (SCI) Institute (www.sci.utah.edu), we're creating datasets of brain electron microscopy (EM) mosaics that are 16 terabytes in size. However, while there's no foreseeable end to the increase in our ability to produce simulation data or record observational data, our ability to use this data in meaningful ways is inhibited by current data analysis capabilities, which already lag far behind. Indeed, as the NIH-NSF Visualization Research Challenges report notes, to effectively understand and make use of the vast amounts of data researchers are producing is one of the greatest scientific challenges of the 21st century.
Visual data analysis involves creating images that convey salient information about underlying data and processes, enabling the detection and validation of expected results while leading to unexpected discoveries in science. This allows for the validation of new theoretical models, provides comparison between models and datasets, enables quantitative and qualitative querying, improves interpretation of data, and facilitates decision making. Scientists can use visual data analysis systems to explore \"what if\" scenarios, define hypotheses, and examine data under multiple perspectives and assumptions. In addition, they can identify connections between numerous attributes and quantitatively assess the reliability of hypotheses. In essence, visual data analysis is an integral part of scientific problem solving and discovery.
As applied to biomedical systems, visualization plays a crucial role in our ability to comprehend large and complex data-data that, in two, three, or more dimensions, convey insight into many diverse biomedical applications, including understanding neural connectivity within the brain, interpreting bioelectric currents within the heart, characterizing white-matter tracts by diffusion tensor imaging, and understanding morphology differences among different genetic mice phenotypes.
Keywords: kaust
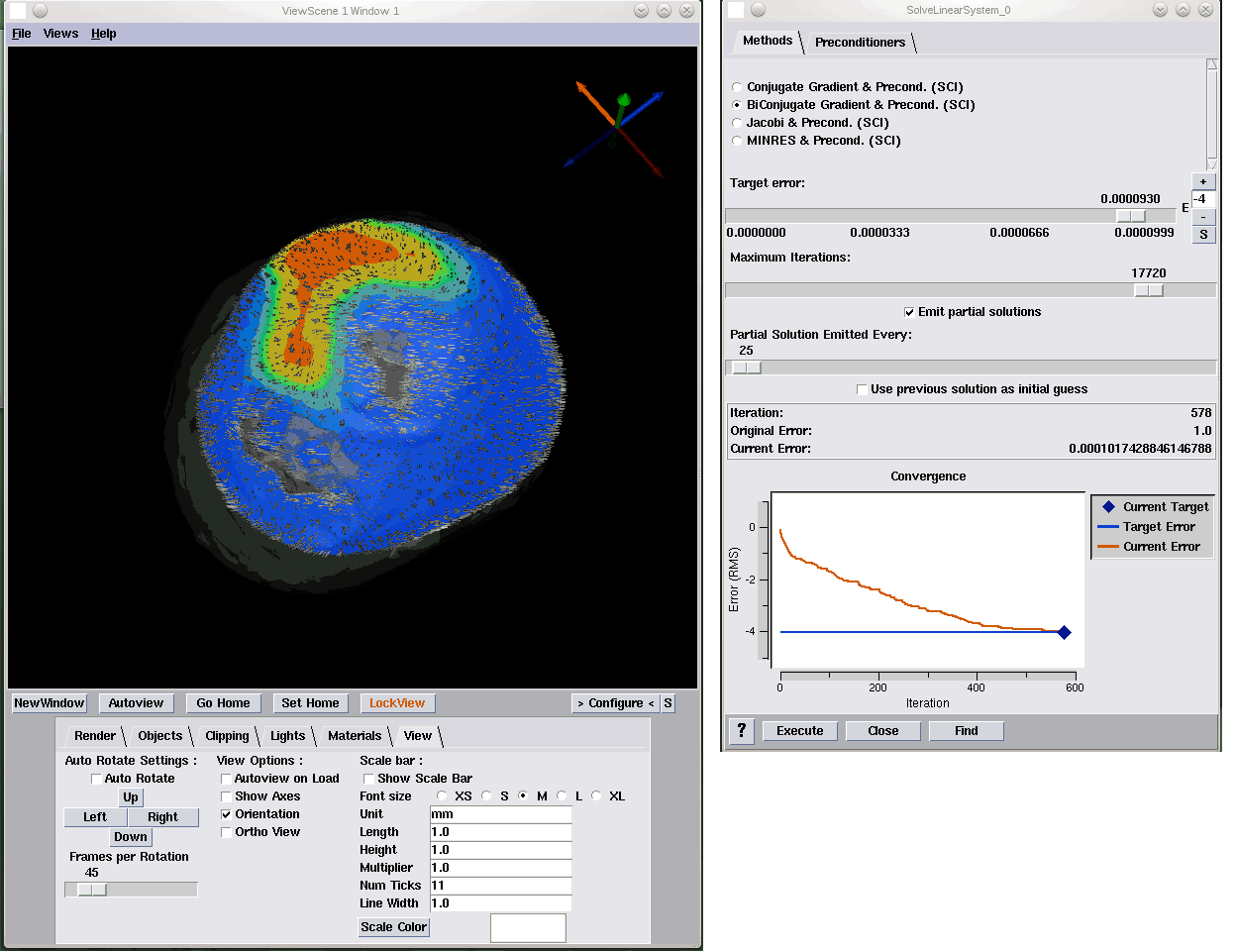
J. Knezevic, R.-P. Mundani, E. Rank, A. Khan, C.R. Johnson.
“Extending the SCIRun Problem Solving Environment to Large-Scale Applications,” In Proceedings of Applied Computing 2012, IADIS, pp. 171--178. October, 2012.
Keywords: scirun
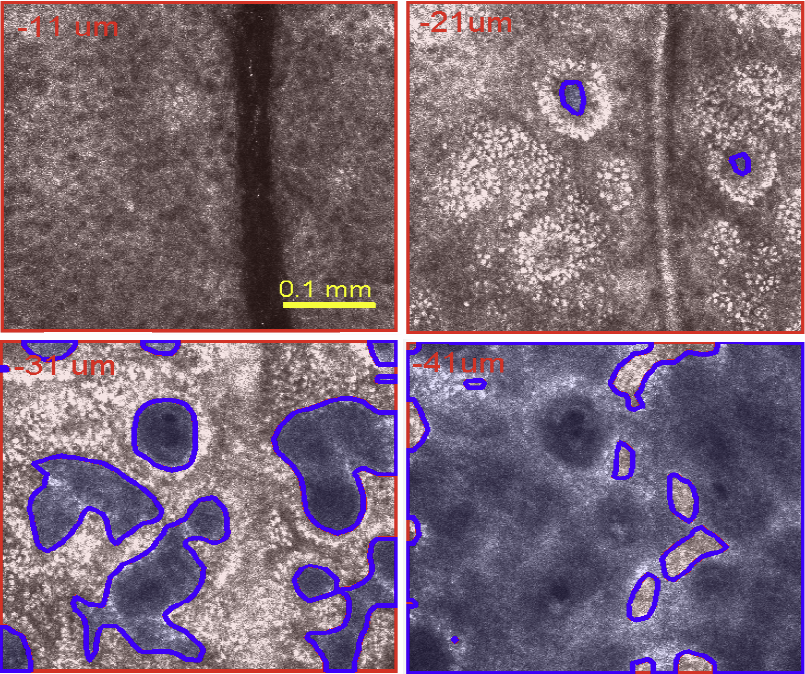
S. Kurugol, M. Rajadhyaksha, J.G. Dy, D.H. Brooks.
“Validation study of automated dermal/epidermal junction localization algorithm in reflectance confocal microscopy images of skin,” In Proceedings of SPIE Photonic Therapeutics and Diagnostics VIII, Vol. 8207, No. 1, pp. 820702-820711. 2012.
DOI: 10.1117/12.909227
PubMed ID: 24376908
PubMed Central ID: PMC3872972
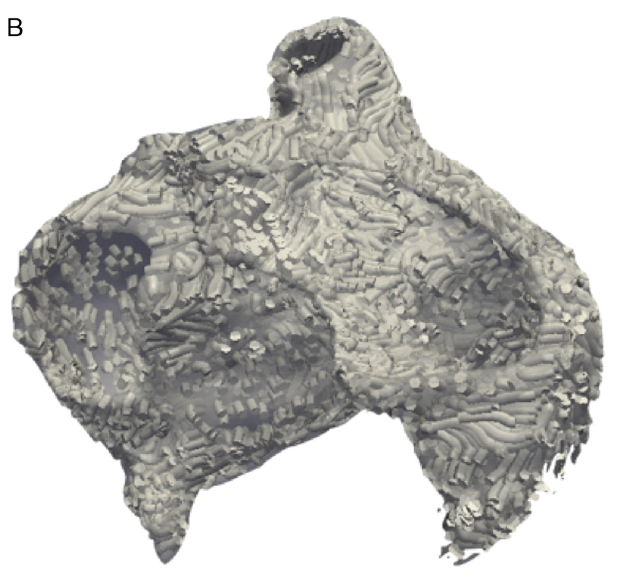
K.S. McDowell, F. Vadakkumpadan, R. Blake, J. Blauer, G. Plank, R.S. MacLeod, N.A. Trayanova.
“Methodology for patient-specific modeling of atrial fibrosis as a substrate for atrial fibrillation,” In Journal of Electrocardiology, Vol. 45, No. 6, pp. 640--645. 2012.
DOI: 10.1016/j.jelectrocard.2012.08.005
PubMed ID: 22999492
PubMed Central ID: PMC3515859
Keywords: Patient-specific modeling, Computational model, Atrial fibrillation, Atrial fibrosis
Q. Meng, J. Hall, H. Rutigliano, X. Zhou, B.R. Sessions, R. Stott, K. Panter, C.J. Davies, R. Ranjan, D. Dosdall, R.S. MacLeod, N. Marrouche, K.L. White, Z. Wang, I.A. Polejaeva.
“30 Generation of Cloned Transgenic Goats with Cardiac Specific Overexpression of Transforming Growth Factor β1,” In Reproduction, Fertility and Development, Vol. 25, No. 1, pp. 162--163. 2012.
DOI: 10.1071/RDv25n1Ab30
Transforming growth factor β1 (TGF-β1) has a potent profibrotic function and is central to signaling cascades involved in interstitial fibrosis, which plays a critical role in the pathobiology of cardiomyopathy and contributes to diastolic and systolic dysfunction. In addition, fibrotic remodeling is responsible for generation of re-entry circuits that promote arrhythmias (Bujak and Frangogiannis 2007 Cardiovasc. Res. 74, 184–195). Due to the small size of the heart, functional electrophysiology of transgenic mice is problematic. Large transgenic animal models have the potential to offer insights into conduction heterogeneity associated with fibrosis and the role of fibrosis in cardiovascular diseases. The goal of this study was to generate transgenic goats overexpressing an active form of TGFβ-1 under control of the cardiac-specific α-myosin heavy chain promoter (α-MHC). A pcDNA3.1DV5-MHC-TGF-β1cys33ser vector was constructed by subcloning the MHC-TGF-β1 fragment from the plasmid pUC-BM20-MHC-TGF-β1 (Nakajima et al. 2000 Circ. Res. 86, 571–579) into the pcDNA3.1D V5 vector. The Neon transfection system was used to electroporate primary goat fetal fibroblasts. After G418 selection and PCR screening, transgenic cells were used for SCNT. Oocytes were collected by slicing ovaries from an abattoir and matured in vitro in an incubator with 5\% CO2 in air. Cumulus cells were removed at 21 to 23 h post-maturation. Oocytes were enucleated by aspirating the first polar body and nearby cytoplasm by micromanipulation in Hepes-buffered SOF medium with 10 µg of cytochalasin B mL–1. Transgenic somatic cells were individually inserted into the perivitelline space and fused with enucleated oocytes using double electrical pulses of 1.8 kV cm–1 (40 µs each). Reconstructed embryos were activated by ionomycin (5 min) and DMAP and cycloheximide (CHX) treatments. Cloned embryos were cultured in G1 medium for 12 to 60 h in vitro and then transferred into synchronized recipient females. Pregnancy was examined by ultrasonography on day 30 post-transfer. A total of 246 cloned embryos were transferred into 14 recipients that resulted in production of 7 kids. The pregnancy rate was higher in the group cultured for 12 h compared with those cultured 36 to 60 h [44.4\% (n = 9) v. 20\% (n = 5)]. The kidding rates per embryo transferred of these 2 groups were 3.8\% (n = 156) and 1.1\% (n = 90), respectively. The PCR results confirmed that all the clones were transgenic. Phenotype characterization [e.g. gene expression, electrocardiogram (ECG), and magnetic resonance imaging (MRI)] is underway. We demonstrated successful production of transgenic goat via SCNT. To our knowledge, this is the first transgenic goat model produced for cardiovascular research.
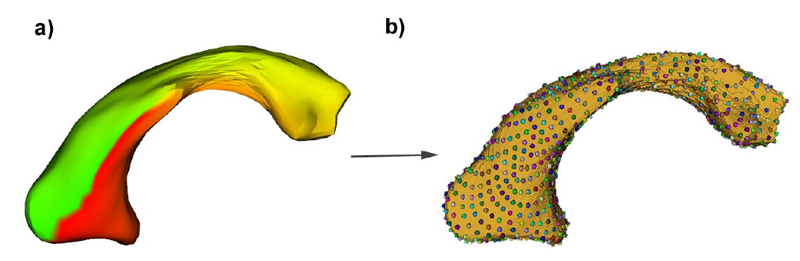
B. Paniagua, L. Bompard, J. Cates, R.T. Whitaker, M. Datar, C. Vachet, M. Styner.
“Combined SPHARM-PDM and entropy-based particle systems shape analysis framework,” In Medical Imaging 2012: Biomedical Applications in Molecular, Structural, and Functional Imaging, SPIE Intl Soc Optical Eng, March, 2012.
DOI: 10.1117/12.911228
PubMed ID: 24027625
PubMed Central ID: PMC3766973
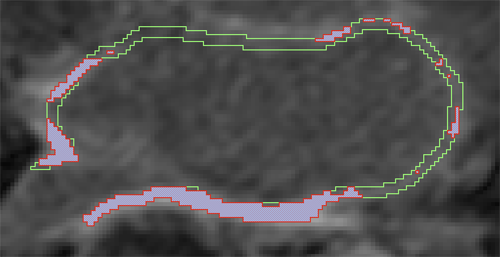
D. Perry, A. Morris, N. Burgon, C. McGann, R.S. MacLeod, J. Cates.
“Automatic classification of scar tissue in late gadolinium enhancement cardiac MRI for the assessment of left-atrial wall injury after radiofrequency ablation,” In SPIE Proceedings, Vol. 8315, pp. (published online). 2012.
DOI: 10.1117/12.910833
PubMed ID: 24236224
PubMed Central ID: PMC3824273
K. Potter, R.M. Kirby, D. Xiu, C.R. Johnson.
“Interactive visualization of probability and cumulative density functions,” In International Journal of Uncertainty Quantification, Vol. 2, No. 4, pp. 397--412. 2012.
DOI: 10.1615/Int.J.UncertaintyQuantification.2012004074
PubMed ID: 23543120
PubMed Central ID: PMC3609671
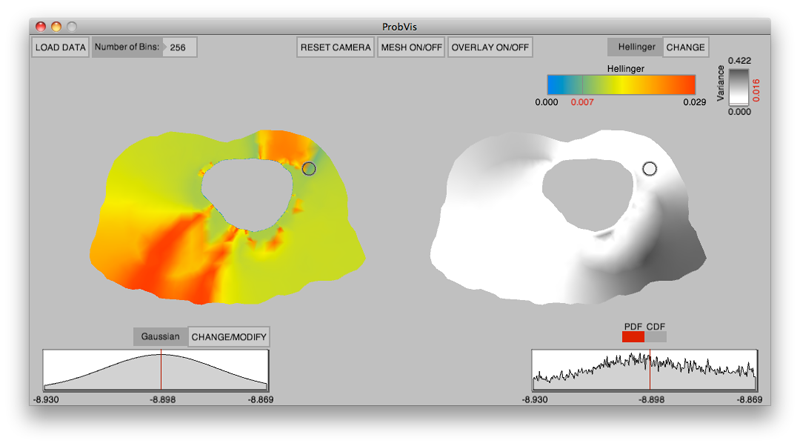
Keywords: visualization, probability density function, cumulative density function, generalized polynomial chaos, stochastic Galerkin methods, stochastic collocation methods
K. Potter, P. Rosen, C.R. Johnson.
“From Quantification to Visualization: A Taxonomy of Uncertainty Visualization Approaches,” In Uncertainty Quantification in Scientific Computing, IFIP Advances in Information and Communication Technology Series, Vol. 377, Edited by Andrew Dienstfrey and Ronald Boisvert, Springer, pp. 226--249. 2012.
DOI: 10.1007/978-3-642-32677-6_15
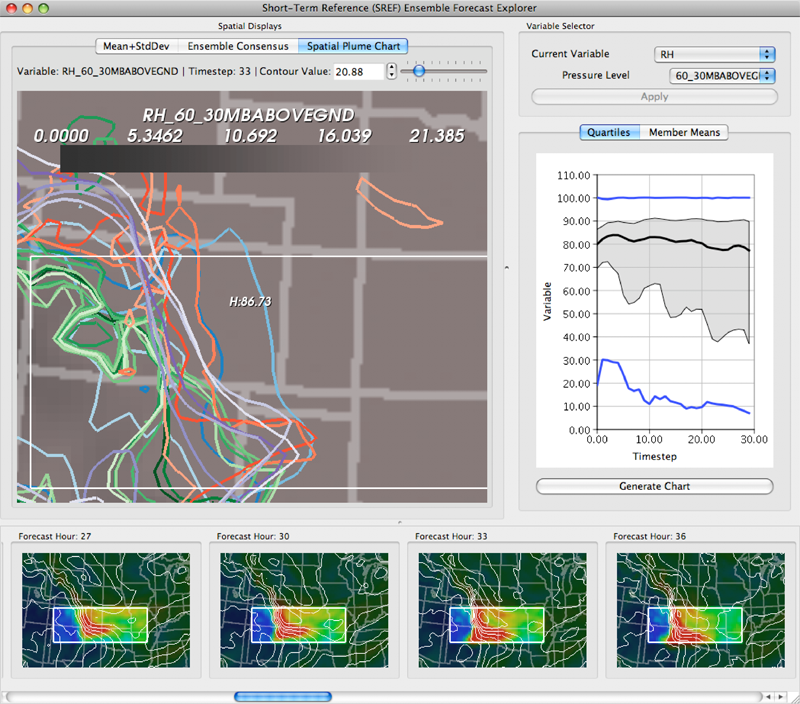
Keywords: scidac, netl, uncertainty visualization
Page 9 of 24