2011
S. Kurugol, J.G. Dy, M. Rajadhyaksha, K.W. Gossage, J. Weissman, D.H. Brooks.
“Semi-automated Algorithm for Localization of Dermal/ Epidermal Junction in Reflectance Confocal Microscopy Images of Human Skin,” In Proceedings of SPIE, Vol. 7904, pp. 79041A-79041A-10. 2011.
DOI: 10.1117/12.875392
PubMed ID: 21709746
PubMed Central ID: PMC3120112
The examination of the dermis/epidermis junction (DEJ) is clinically important for skin cancer diagnosis. Reflectance confocal microscopy (RCM) is an emerging tool for detection of skin cancers in vivo. However, visual localization of the DEJ in RCM images, with high accuracy and repeatability, is challenging, especially in fair skin, due to low contrast, heterogeneous structure and high inter- and intra-subject variability. We recently proposed a semi-automated algorithm to localize the DEJ in z-stacks of RCM images of fair skin, based on feature segmentation and classification. Here we extend the algorithm to dark skin. The extended algorithm first decides the skin type and then applies the appropriate DEJ localization method. In dark skin, strong backscatter from the pigment melanin causes the basal cells above the DEJ to appear with high contrast. To locate those high contrast regions, the algorithm operates on small tiles (regions) and finds the peaks of the smoothed average intensity depth profile of each tile. However, for some tiles, due to heterogeneity, multiple peaks in the depth profile exist and the strongest peak might not be the basal layer peak. To select the correct peak, basal cells are represented with a vector of texture features. The peak with most similar features to this feature vector is selected. The results show that the algorithm detected the skin types correctly for all 17 stacks tested (8 fair, 9 dark). The DEJ detection algorithm achieved an average distance from the ground truth DEJ surface of around 4.7μm for dark skin and around 7-14μm for fair skin.
Keywords: confocal reflectance microscopy, image analysis, skin, classification
S. Kurugol, J.G. Dy, D.H. Brooks, M. Rajadhyaksha.
“Pilot study of semiautomated localization of the dermal/epidermal junction in reflectance confocal microscopy images of skin,” In Journal of biomedical optics, Vol. 16, No. 3, International Society for Optics and Photonics, pp. 036005--036005. 2011.
DOI: 10.1117/1.3549740
Reflectance confocal microscopy (RCM) continues to be translated toward the detection of skin cancers in vivo. Automated image analysis may help clinicians and accelerate clinical acceptance of RCM. For screening and diagnosis of cancer, the dermal/epidermal junction (DEJ), at which melanomas and basal cell carcinomas originate, is an important feature in skin. In RCM images, the DEJ is marked by optically subtle changes and features and is difficult to detect purely by visual examination. Challenges for automation of DEJ detection include heterogeneity of skin tissue, high inter-, intra-subject variability, and low optical contrast. To cope with these challenges, we propose a semiautomated hybrid sequence segmentation/classification algorithm that partitions z-stacks of tiles into homogeneous segments by fitting a model of skin layer dynamics and then classifies tile segments as epidermis, dermis, or transitional DEJ region using texture features. We evaluate two different training scenarios: 1. training and testing on portions of the same stack; 2. training on one labeled stack and testing on one from a different subject with similar skin type. Initial results demonstrate the detectability of the DEJ in both scenarios with epidermis/dermis misclassification rates smaller than 10% and average distance from the expert labeled boundaries around 8.5 μm.
M. Leeser, D. Yablonski, D.H. Brooks, L.S. King.
“The Challenges of Writing Portable, Correct and High Performance Libraries for GPUs,” In Computer Architecture News, Vol. 39, No. 4, pp. 2--7. 2011.
DOI: 10.1145/2082156.2082158
Graphics Processing Units (GPUs) are widely used to accelerate scientific applications. Many successes have been reported with speedups of two or three orders of magnitude over serial implementations of the same algorithms. These speedups typically pertain to a specific implementation with fixed parameters mapped to a specific hardware implementation. The implementations are not designed to be easily ported to other GPUs, even from the same manufacturer. When target hardware changes, the application must be re-optimized.
In this paper we address a different problem. We aim to deliver working, efficient GPU code in a library that is downloaded and run by many different users. The issue is to deliver efficiency independent of the individual user parameters and without a priori knowledge of the hardware the user will employ. This problem requires a different set of tradeoffs than finding the best runtime for a single solution. Solutions must be adaptable to a range of different parameters both to solve users' problems and to make the best use of the target hardware.
Another issue is the integration of GPUs into a Problem Solving Environment (PSE) where the use of a GPU is almost invisible from the perspective of the user. Ease of use and smooth interactions with the existing user interface are important to our approach. We illustrate our solution with the incorporation of GPU processing into the Scientific Computing Institute (SCI)Run Biomedical PSE developed at the University of Utah. SCIRun allows scientists to interactively construct many different types of biomedical simulations. We use this environment to demonstrate the effectiveness of the GPU by accelerating time consuming algorithms in the scientist's simulations. Specifically we target the linear solver module, including Conjugate Gradient, Jacobi and MinRes solvers for sparse matrices.
R.S. MacLeod, J.J.E. Blauer.
“Atrial Fibrillation,” In Multimodal Cardiovascular Imaging: Principles and Clinical Applications, Ch. 25, Edited by O. Pahlm and G. Wagner, McGraw Hill, 2011.
ISBN: 0071613463
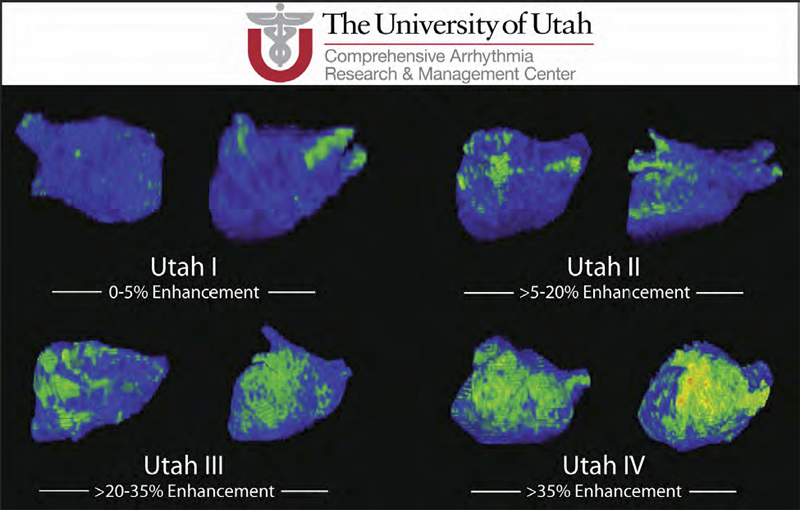
Atrial fibrillation (AF) is the most common form of cardiac arrhythmia so that a review of the role imaging in AF is a natural topic to include in this book. Further motivation comes from the fact that the treatment of AF probably includes more different forms of imaging, often merged or combined in a variety of ways, than perhaps any other clinical intervention. A typical clinical electrophysiology lab for the treatment of AF usually contains no less than 6 and often more than 8 individual monitors, each rendering some form of image based information about the patient undergoing therapy. There is naturally great motivation to merge different images and different imaging modalities in the setting of AF but also very challenging because of a host of factors related to the small size, extremely thin walls, the large natural variation in atrial shape, and the fact that fibrillation is occurring so that atrial shape is changing rapidly and irregularly. Thus, the use of multimodal imaging has recently become a very active and challenging area of image processing and analysis research and development, driven by an enormous clinical need to understand and treat a disease that affects some 5 million Americans alone, a number that is predicted to increase to almost 16 million by 2050.
In this chapter we attempt to provide an overview of the large variety of imaging modalities and uses in the management and understanding of atrial fibrillation, with special emphasis on the most novel applications of magnetic resonance imaging (MRI) technology. To provide clinical and biomedical motivation, we outline the basics of the disease together with some contemporary hypotheses about its etiology and management. We then describe briefly the imaging modalities in common use in the management and research of AF, then focus on the use or MRI for all phases of the management of patients with AF and indicate some of the major engineering challenges that can motivate further progress.
Keywords: ablation, carma, cvrti, 5P41-RR012553-10
M.Q. Madrigal, G. Tadmor, G. C. Cano, D.H. Brooks.
“Low-Order 4D Dynamical Modeling of Heart Motion Under Respiration,” In Proceeding of the IEEE International Symposium on Biomedical Imaging: from nano to macro, pp. 1326--1329. 2011.
PubMed ID: 21927642
PubMed Central ID: PMC3172964
This work is motivated by the limitations of current techniques to visualize the heart as it moves under contraction and respiration during interventional procedures such as ablation of atrial fibrillation. Our long-term goal is to integrate high resolution models routinely obtained from pre-procedure imaging (here, via MRI) with the low resolution, sparse, images, along with a few scalar measurements such as ECG, which are feasible during the real-time procedure. A key ingredient to facilitate this integration is the extraction from the pre-procedure model of an individualized, low complexity, dynamic model of the moving and beating heart. This is the immediate goal we address here. Our approach stems from work on distributed parameter dynamical systems and uses a combination of truncated basis expansions to obtain the requisite four dimensional low order model. The method's potential is illustrated not only by modeling results but also by estimation of an arbitrary slice from the parameterized model.
C.J. McGann, E.G. Kholmovski, J.J. Blauer, S. Vijayakumar, T.S. Haslam, J.E. Cates, E.V. DiBella, N.S. Burgon, B. Wilson, A.J. Alexander, M.W. Prastawa, M. Daccarett, G. Vergara, N.W. Akoum, D.L. Parker, R.S. MacLeod, N.F. Marrouche.
“Dark Regions of No-Reflow on Late Gadolinium Enhancement Magnetic Resonance Imaging Result in Scar Formation After Atrial Fibrillation Ablation,” In Journal of the American College of Cardiology, Vol. 58, No. 2, pp. 177--185. 2011.
DOI: 10.1016/j.jacc.2011.04.008
PubMed ID: 21718914
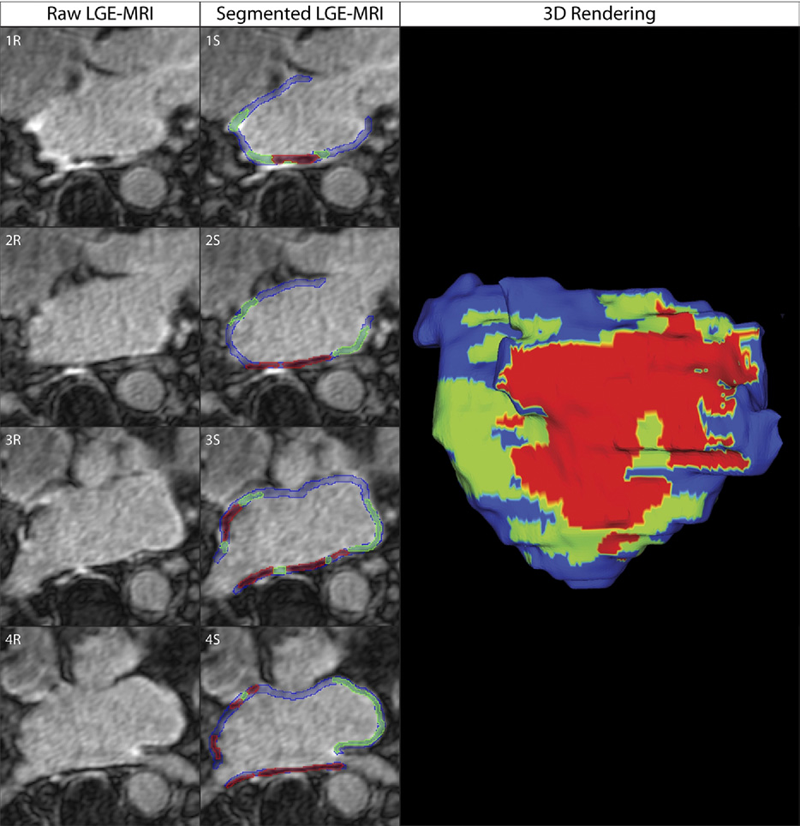
Objectives: The aim of this study was to assess acute ablation injuries seen on late gadolinium enhancement (LGE) magnetic resonance imaging (MRI) immediately post-ablation (IPA) and the association with permanent scar 3 months post-ablation (3moPA).
Background: Success rates for atrial fibrillation catheter ablation vary significantly, in part because of limited information about the location, extent, and permanence of ablation injury at the time of procedure. Although the amount of scar on LGE MRI months after ablation correlates with procedure outcomes, early imaging predictors of scar remain elusive.
Methods: Thirty-seven patients presenting for atrial fibrillation ablation underwent high-resolution MRI with a 3-dimensional LGE sequence before ablation, IPA, and 3moPA using a 3-T scanner. The acute left atrial wall injuries on IPA scans were categorized as hyperenhancing (HE) or nonenhancing (NE) and compared with scar 3moPA.
Results: Heterogeneous injuries with HE and NE regions were identified in all patients. Dark NE regions in the left atrial wall on LGE MRI demonstrate findings similar to the \"no-reflow\" phenomenon. Although the left atrial wall showed similar amounts of HE, NE, and normal tissue IPA (37.7 ± 13\%, 34.3 ± 14\%, and 28.0 ± 11\%, respectively; p = NS), registration of IPA injuries with 3moPA scarring demonstrated that 59.0 ± 19\% of scar resulted from NE tissue, 30.6 ± 15\% from HE tissue, and 10.4 ± 5\% from tissue identified as normal. Paired t-test comparisons were all statistically significant among NE, HE, and normal tissue types (p less than 0.001). Arrhythmia recurrence at 1-year follow-up correlated with the degree of wall enhancement 3moPA (p = 0.02).
Conclusion: Radiofrequency ablation results in heterogeneous injury on LGE MRI with both HE and NE wall lesions. The NE lesions demonstrate no-reflow characteristics and reveal a better predictor of final scar at 3 months. Scar correlates with procedure outcomes, further highlighting the importance of early scar prediction. (J Am Coll Cardiol 2011;58:177–85) © 2011 by the American College of Cardiology Foundation
T.A. Quinn, S. Granite, M.A. Allessie, C. Antzelevitch, C. Bollensdorff, G. Bub, R.A.B. Burton, E. Cerbai, P.S. Chen, M. Delmar, D. DiFrancesco, Y.E. Earm, I.R. Efimov, M. Egger, E. Entcheva, M. Fink, R. Fischmeister, M.R. Franz, A. Garny, W.R. Giles, T. Hannes, S.E. Harding, P.J. Hunter, s, G. Iribe, J. Jalife, C.R. Johnson, R.S. Kass, I. Kodama, G. Koren, P. Lord, V.S. Markhasin, S. Matsuoka, A.D. McCulloch, G.R. Mirams, G.E. Morley, S. Nattel, D. Noble, S.P. Olesen, A.V. Panfilov, N.A. Trayanova, U. Ravens, S. Richard, D.S. Rosenbaum, Y. Rudy, F. Sachs, F.B. Sachse, D.A. Saint, U. Schotten, O. Solovyova, P. Taggart, L. Tung, A. Varrò, P.G. Volders, K. Wang, J.N. Weiss, E. Wettwer, E. White, R. Wilders, R.L. Winslow, P. Kohl.
“Minimum Information about a Cardiac Electrophysiology Experiment (MICEE): Standardised reporting for model reproducibility, interoperability, and data sharing,” In Progress in Biophysics and Molecular Biology, Vol. 107, No. 1, Elsevier, pp. 4--10. October, 2011.
DOI: 10.1016/j.pbiomolbio.2011.07.001
PubMed Central ID: PMC3190048
Cardiac experimental electrophysiology is in need of a well-defined Minimum Information Standard for recording, annotating, and reporting experimental data. As a step toward establishing this, we present a draft standard, called Minimum Information about a Cardiac Electrophysiology Experiment (MICEE). The ultimate goal is to develop a useful tool for cardiac electrophysiologists which facilitates and improves dissemination of the minimum information necessary for reproduction of cardiac electrophysiology research, allowing for easier comparison and utilisation of findings by others. It is hoped that this will enhance the integration of individual results into experimental, computational, and conceptual models. In its present form, this draft is intended for assessment and development by the research community. We invite the reader to join this effort, and, if deemed productive, implement the Minimum Information about a Cardiac Electrophysiology Experiment standard in their own work.
Keywords: Minimum Information Standard; Cardiac electrophysiology; Data sharing; Reproducibility; Integration; Computational modelling
P. Rosen, V. Popescu.
“An Evaluation of 3-D Scene Exploration Using a Multiperspective Image Framework,” In The Visual Computer, Vol. 27, No. 6-8, Springer-Verlag New York, Inc., pp. 623--632. 2011.
DOI: 10.1007/s00371-011-0599-2
PubMed ID: 22661796
PubMed Central ID: PMC3364594
Multiperspective images (MPIs) show more than what is visible from a single viewpoint and are a promising approach for alleviating the problem of occlusions. We present a comprehensive user study that investigates the effectiveness of MPIs for 3-D scene exploration. A total of 47 subjects performed searching, counting, and spatial orientation tasks using both conventional and multiperspective images. We use a flexible MPI framework that allows trading off disocclusion power for image simplicity. The framework also allows rendering MPI images at interactive rates, which enables investigating interactive navigation and dynamic 3-D scenes. The results of our experiments show that MPIs can greatly outperform conventional images. For searching, subjects performed on average 28% faster using an MPI. For counting, accuracy was on average 91% using MPIs as compared to 42% for conventional images.
Keywords: Interactive 3-D scene exploration, Navigation, Occlusions, User study, Visual interfaces
R. Samuel, H.J. Sant, F. Jiao, C.R. Johnson, B.K. Gale.
“Microfluidic laminate-based phantom for diffusion tensor-magnetic resonance imaging,” In Journal of Micromech. Microeng., Vol. 21, pp. 095027--095038. 2011.
DOI: 10.1088/0960-1317/21/9/095027
D.J. Swenson, S.E. Geneser, J.G. Stinstra, R.M. Kirby, R.S. MacLeod.
“Cardiac Position Sensitivity Study in the Electrocardiographic Forward Problem Using Stochastic Collocation and Boundary Element Methods,” In Annals of Biomedical Engineering, Vol. 39, No. 12, pp. 2900--2910. 2011.
DOI: 10.1007/s10439-011-0391-5
PubMed ID: 21909818
PubMed Central ID: PMC336204
The electrocardiogram (ECG) is ubiquitously employed as a diagnostic and monitoring tool for patients experiencing cardiac distress and/or disease. It is widely known that changes in heart position resulting from, for example, posture of the patient (sitting, standing, lying) and respiration significantly affect the body-surface potentials; however, few studies have quantitatively and systematically evaluated the effects of heart displacement on the ECG. The goal of this study was to evaluate the impact of positional changes of the heart on the ECG in the specific clinical setting of myocardial ischemia. To carry out the necessary comprehensive sensitivity analysis, we applied a relatively novel and highly efficient statistical approach, the generalized polynomial chaos-stochastic collocation method, to a boundary element formulation of the electrocardiographic forward problem, and we drove these simulations with measured epicardial potentials from whole-heart experiments. Results of the analysis identified regions on the body-surface where the potentials were especially sensitive to realistic heart motion. The standard deviation (STD) of ST-segment voltage changes caused by the apex of a normal heart, swinging forward and backward or side-to-side was approximately 0.2 mV. Variations were even larger, 0.3 mV, for a heart exhibiting elevated ischemic potentials. These variations could be large enough to mask or to mimic signs of ischemia in the ECG. Our results suggest possible modifications to ECG protocols that could reduce the diagnostic error related to postural changes in patients possibly suffering from myocardial ischemia.
G. Tamm, A. Schiewe, J. Krüger.
“ZAPP – A management framework for distributed visualization systems,” In Proceedings of CGVCVIP 2011 : IADIS International Conference on Computer Graphics, Visualization, Computer Vision And Image Processing, pp. (accepted). 2011.
J.D. Tate, J.G. Stinstra, T. Pilcher, A. Poursaid, E. Saarel, R.S. MacLeod.
“Measuring Defibrillator Surface Potentials for Simulation Verification,” In Proceedings of the IEEE Engineering in Medicine and Biology Society EMBS 33rd Annual International Conference, pp. 239 - 242. 2011.
ISSN: 1557-170X
DOI: 10.1109/IEMBS.2011.6090045
PubMed ID: 22254294
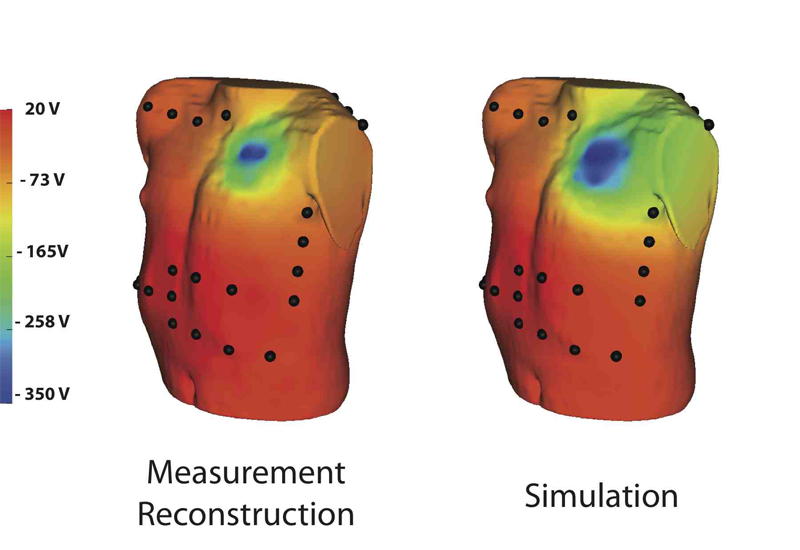
Though implantable cardioverter defibrillators (ICDs) are increasing in use in both adults and children, little progress has been devoted to optimizing device and electrode placement. To facilitate effective ICD placement, especially in pediatric cases, we have developed a predictive model that evaluates the efficacy of a delivered shock. We have also developed an experimental validation approach based on measurements from clinical cases. The approach involves obtaining body surface potential maps of ICD discharges during implantation surgery using a limited lead selection and body surface estimation algorithm. Comparison of the simulated and measured potentials yielded very similar patterns and a typical correlation greater than 0.93, suggesting that the predictive simulation generates realistic potential values. This validation approach provides confidence in application of the simulation pipeline and offers areas to focus future improvements.
Keywords: Electric potential, Electric shock, Electrodes;Estimation, Lead;Surface reconstruction, Torso, Algorithms, Body Surface Potential Mapping, Computer Simulation, Defibrillators, Implantable, Humans, Models, Cardiovascular, Therapy, Computer-Assisted
J.D. Tate, J.G. Stinstra, T.A. Pilcher, R.S. MacLeod.
“Measurement of Defibrillator Surface Potentials for Simulation Verification,” In Computing in Cardiology, In 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, pp. 853--856. Aug, 2011.
DOI: 10.1109/iembs.2011.6090045
G.R. Vergara, S. Vijayakumar, E.G. Kholmovski, J.J. Blauer, M.A. Guttman, C. Gloschat, G. Payne, K. Vij, N.W. Akoum, M. Daccarett, C.J. McGann, R.S. Macleod, N.F. Marrouche.
“Real-time magnetic resonance imaging-guided radiofrequency atrial ablation and visualization of lesion formation at 3 Tesla,” In Heart Rhythm, Vol. 8, No. 2, pp. 295--303. 2011.
PubMed ID: 21034854
D. Wang, R.M. Kirby, C.R. Johnson.
“Finite Element Based Discretization and Regularization Strategies for 3D Inverse Electrocardiography,” In IEEE Transactions for Biomedical Engineering, Vol. 58, No. 6, pp. 1827--1838. 2011.
PubMed ID: 21382763
PubMed Central ID: PMC3109267
D. Wang, R.M. Kirby, R.S. Macleod, C.R. Johnson.
“An optimization framework for inversely estimating myocardial transmembrane potentials and localizing ischemia,” In Proceedings of the International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), pp. 1680--1683. 2011.
DOI: 10.1109/IEMBS.2011.6090483
PubMed ID: 22254648
PubMed Central ID: PMC3336368
2010
J.R. Anderson, B.C. Grimm, S. Mohammed, B.W. Jones, T. Tasdizen, J. Spaltenstein, P. Koshevoy, R.T. Whitaker, R.E. Marc.
“The Viking Viewer: Scalable Multiuser Annotation and Summarization of Large Volume Datasets,” In Journal of Microscopy, Vol. 241, No. 1, pp. 13--28. 2010.
DOI: 10.1111/j.1365-2818.2010.03402.x
T.J. Badger, M. Daccarett, N.W. Akoum, Y.A. Adjei-Poku, N.S. Burgon, T.S. Haslam, S. Kalvaitis, S. Kuppahally, G. Vergara, L. McMullen, P.A. Anderson, E. Kholmovski, R.S. Macleod, N.F. Marrouche.
“Evaluation of left atrial lesions after initial and repeat atrial fibrillation ablation: lessons learned from delayed-enhancement MRI in repeat ablation procedures,” In Circulation. Arrhythmia and Electrophysiology, Vol. 3, No. 3, pp. 249--259. 2010.
PubMed ID: 20335558
J.J.E. Blauer, J. Cates, C.J. McGann, E.G. Kholmovski, A. Alexander, M.W. Prastawa, S. Joshi, N.F. Marrouche, R.S. MacLeod.
“MRI Based Injury Characterization Immediately Following Ablation of Atrial Fibrillation,” In Computing in Cardiology, Vol. 37, pp. 165--168. 2010.
ISSN: 0276−6574
J.R. Bronson, J.A. Levine, R.T. Whitaker.
“Particle Systems for Adaptive, Isotropic Meshing of CAD Models,” In Proceedings of the 19th International Meshing Roundtable, Note: Awarded Best Paper, Springer, pp. 279-296. 2010.
DOI: 10.1007/978-3-642-15414-0_17
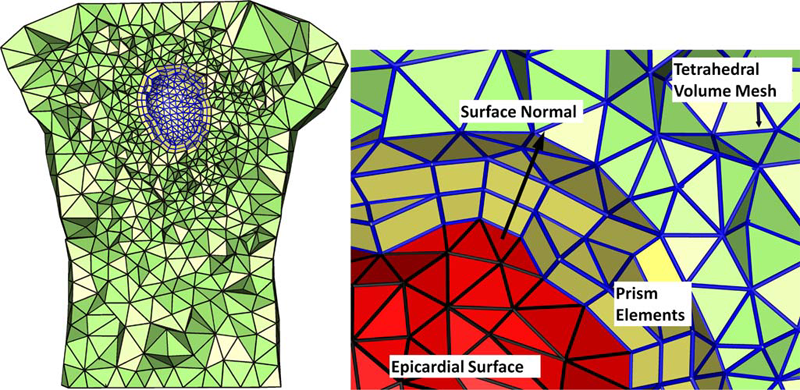
We present a particle-based approach for generating adaptive triangular surface and tetrahedral volume meshes from CAD models. Input shapes are treated as a collection of smooth, parametric surface patches that can meet non-smoothly on boundaries. Our approach uses a hierarchical sampling scheme that places particles on features in order of increasing dimensionality. These particles reach a good distribution by minimizing an energy computed in 3D world space, with movements occurring in the parametric space of each surface patch.
Rather than using a pre-computed measure of feature size, our system automatically adapts to both curvature as well as a notion of topological separation. It also enforces a measure of smoothness on these constraints to construct a sizing field that acts as a proxy to piecewise-smooth feature size. We evaluate our technique with comparisons against other popular triangular meshing techniques for this domain.
Page 11 of 24