SCI Publications
2011
T. Ize, C.D. Hansen.
“RTSAH Traversal Order for Occlusion Rays,” In Computer Graphics Forum, Vol. 30, No. 2, Wiley-Blackwell, pp. 297--305. April, 2011.
DOI: 10.1111/j.1467-8659.2011.01861.x

T. Ize, C. Brownlee, C.D. Hansen.
“Real-Time Ray Tracer for Visualizing Massive Models on a Cluster,” In Proceedings of the 2011 Eurographics Symposium on Parallel Graphics and Visualization, pp. 61--69. 2011.

S. Jadhav, H. Bhatia, P.-T. Bremer, J.A. Levine, L.G. Nonato, V. Pascucci.
“Consistent Approximation of Local Flow Behavior for 2D Vector Fields,” In Mathematics and Visualization, Springer, pp. 141--159. Nov, 2011.
DOI: 10.1007/978-3-642-23175-9 10
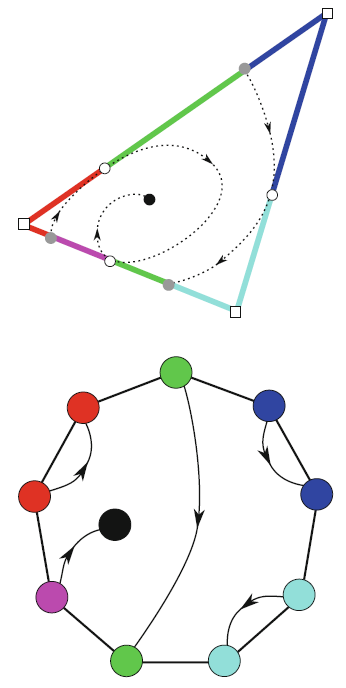
Typically, vector fields are stored as a set of sample vectors at discrete locations. Vector values at unsampled points are defined by interpolating some subset of the known sample values. In this work, we consider two-dimensional domains represented as triangular meshes with samples at all vertices, and vector values on the interior of each triangle are computed by piecewise linear interpolation.
Many of the commonly used techniques for studying properties of the vector field require integration techniques that are prone to inconsistent results. Analysis based on such inconsistent results may lead to incorrect conclusions about the data. For example, vector field visualization techniques integrate the paths of massless particles (streamlines) in the flow or advect a texture using line integral convolution (LIC). Techniques like computation of the topological skeleton of a vector field, require integrating separatrices, which are streamlines that asymptotically bound regions where the flow behaves differently. Since these integrations may lead to compound numerical errors, the computed streamlines may intersect, violating some of their fundamental properties such as being pairwise disjoint. Detecting these computational artifacts to allow further analysis to proceed normally remains a significant challenge.
J. Jakeman, R. Archibald, D. Xiu.
“Characterization of Discontinuities in High-dimensional Stochastic Problmes on Adaptive Sparse Grids,” In Journal of Computational Physics, Vol. 230, No. 10, pp. 3977--3997. 2011.
DOI: 10.1016/j.jcp.2011.02.022
Keywords: Adaptive sparse grids, Stochastic partial differential equations, Multivariate discontinuity detection, Generalized polynomial chaos method, High-dimensional approximation
F. Jiao, Y. Gur, C.R. Johnson, S. Joshi.
“Detection of crossing white matter fibers with high-order tensors and rank-k decompositions,” In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI 2011), Lecture Notes in Computer Science (LNCS), Vol. 6801, pp. 538--549. 2011.
DOI: 10.1007/978-3-642-22092-0_44
PubMed Central ID: PMC3327305
P.K. Jimack, R.M. Kirby.
“Towards the Development on an h-p-Refinement Strategy Based Upon Error Estimate Sensitivity,” In Computers and Fluids, Vol. 46, No. 1, pp. 277--281. 2011.
The use of (a posteriori) error estimates is a fundamental tool in the application of adaptive numerical methods across a range of fluid flow problems. Such estimates are incomplete however, in that they do not necessarily indicate where to refine in order to achieve the most impact on the error, nor what type of refinement (for example h-refinement or p-refinement) will be best. This paper extends preliminary work of the authors (Comm Comp Phys, 2010;7:631–8), which uses adjoint-based sensitivity estimates in order to address these questions, to include application with p-refinement to arbitrary order and the use of practical a posteriori estimates. Results are presented which demonstrate that the proposed approach can guide both the h-refinement and the p-refinement processes, to yield improvements in the adaptive strategy compared to the use of more orthodox criteria.
E. Jurrus, S. Watanabe, R. Guily, A.R.C. Paiva, M.H. Ellisman, E.M. Jorgensen, T. Tasdizen.
“Semi-automated Neuron Boundary Detection and Slice Traversal Algorithm for Segmentation of Neurons from Electron Microscopy Images,” In Microscopic Image Analysis with Applications in Biology (MIAAB) Workshop, 2011.
Y. Keller, Y. Gur.
“A Diffusion Approach to Network Localization,” In IEEE Transactions on Signal Processing, Vol. 59, No. 6, pp. 2642--2654. 2011.
DOI: 10.1109/TSP.2011.2122261
D. Keyes, V. Taylor, T. Hey, S. Feldman, G. Allen, P. Colella, P. Cummings, F. Darema, J. Dongarra, T. Dunning, M. Ellisman, I. Foster, W. Gropp, C.R. Johnson, C. Kamath, R. Madduri, M. Mascagni, S.G. Parker, P. Raghavan, A. Trefethen, S. Valcourt, A. Patra, F. Choudhury, C. Cooper, P. McCartney, M. Parashar, T. Russell, B. Schneider, J. Schopf, N. Sharp.
“Advisory Committee for CyberInfrastructure Task Force on Software for Science and Engineering,” Note: NSF Report, 2011.
The Software for Science and Engineering (SSE) Task Force commenced in June 2009 with a charge that consisted of the following three elements:
Identify specific needs and opportunities across the spectrum of scientific software infrastructure. Characterize the specific needs and analyze technical gaps and opportunities for NSF to meet those needs through individual and systemic approaches. Design responsive approaches. Develop initiatives and programs led (or co-led) by NSF to grow, develop, and sustain the software infrastructure needed to support NSF’s mission of transformative research and innovation leading to scientific leadership and technological competitiveness. Address issues of institutional barriers. Anticipate, analyze and address both institutional and exogenous barriers to NSF’s promotion of such an infrastructure.The SSE Task Force members participated in bi-weekly telecons to address the given charge. The telecons often included additional distinguished members of the scientific community beyond the task force membership engaged in software issues, as well as personnel from federal agencies outside of NSF who manage software programs. It was quickly acknowledged that a number of reports loosely and tightly related to SSE existed and should be leveraged. By September 2009, the task formed had formed three subcommittees focused on the following topics: (1) compute-intensive science, (2) data-intensive science, and (3) software evolution.
S.H. Kim, V. Fonov, J. Piven, J. Gilmore, C. Vachet, G. Gerig, D.L. Collins, M. Styner.
“Spatial Intensity Prior Correction for Tissue Segmentation in the Developing human Brain,” In Proceedings of IEEE ISBI 2011, pp. 2049--2052. 2011.
DOI: 10.1109/ISBI.2011.5872815
R.M. Kirby, B. Cockburn, S.J. Sherwin.
“To CG or to HDG: A Comparative Study,” In Journal of Scientific Computing, Note: published online, 2011.
DOI: 10.1007/s10915-011-9501-7
Hybridization through the border of the elements (hybrid unknowns) combined with a Schur complement procedure (often called static condensation in the context of continuous Galerkin linear elasticity computations) has in various forms been advocated in the mathematical and engineering literature as a means of accomplishing domain decomposition, of obtaining increased accuracy and convergence results, and of algorithm optimization. Recent work on the hybridization of mixed methods, and in particular of the discontinuous Galerkin (DG) method, holds the promise of capitalizing on the three aforementioned properties; in particular, of generating a numerical scheme that is discontinuous in both the primary and flux variables, is locally conservative, and is computationally competitive with traditional continuous Galerkin (CG) approaches. In this paper we present both implementation and optimization strategies for the Hybridizable Discontinuous Galerkin (HDG) method applied to two dimensional elliptic operators. We implement our HDG approach within a spectral/hp element framework so that comparisons can be done between HDG and the traditional CG approach.
We demonstrate that the HDG approach generates a global trace space system for the unknown that although larger in rank than the traditional static condensation system in CG, has significantly smaller bandwidth at moderate polynomial orders. We show that if one ignores set-up costs, above approximately fourth-degree polynomial expansions on triangles and quadrilaterals the HDG method can be made to be as efficient as the CG approach, making it competitive for time-dependent problems even before taking into consideration other properties of DG schemes such as their superconvergence properties and their ability to handle hp-adaptivity.
R.C. Knickmeyer, C. Kang, S. Woolson, K.J. Smith, R.M. Hamer, W. Lin, G. Gerig, M. Styner, J.H. Gilmore.
“Twin-Singleton Differences in Neonatal Brain Structure,” In Twin Research and Human Genetics, Vol. 14, No. 3, pp. 268--276. 2011.
ISSN: 1832-4274
DOI: 10.1375/twin.14.3.268
A. Knoll, S. Thelen, I. Wald, C.D. Hansen, H. Hagen, M.E. Papka.
“Full-Resolution Interactive CPU Volume Rendering with Coherent BVH Traversal,” In Proceedings of IEEE Pacific Visualization 2011, pp. 3--10. 2011.
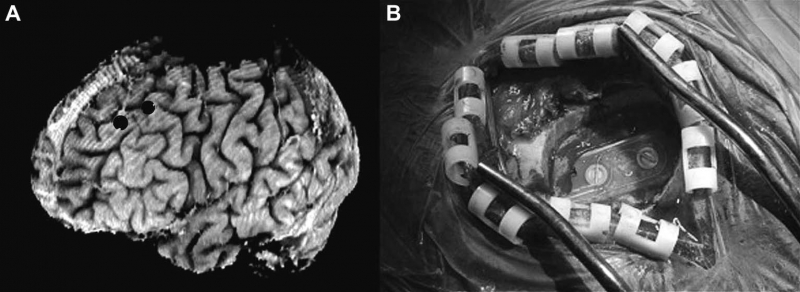
B.H. Kopell, J. Halverson, C.R. Butson, M. Dickinson, J. Bobholz, H. Harsch, C. Rainey, D. Kondziolka, R. Howland, E. Eskandar, K.C. Evans, D.D. Dougherty.
“Epidural cortical stimulation of the left dorsolateral prefrontal cortex for refractory major depressive disorder,” In Neurosurgery, Vol. 69, No. 5, pp. 1015--1029. November, 2011.
ISSN: 1524-4040
DOI: 10.1227/NEU.0b013e318229cfcd
S. Kumar, V. Vishwanath, P. Carns, B. Summa, G. Scorzelli, V. Pascucci, R. Ross, J. Chen, H. Kolla, R. Grout.
“PIDX: Efficient Parallel I/O for Multi-resolution Multi-dimensional Scientific Datasets,” In Proceedings of The IEEE International Conference on Cluster Computing, pp. 103--111. September, 2011.
Z. Leng, J.R. Korenberg, B. Roysam, T. Tasdizen.
“A rapid 2-D centerline extraction method based on tensor voting,” In 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1000--1003. 2011.
DOI: 10.1109/ISBI.2011.5872570
A. Lex, H. Schulz, M. Streit, C. Partl, D. Schmalstieg.
“VisBricks: Multiform Visualization of Large, Inhomogeneous Data,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '11), Vol. 17, No. 12, 2011.
Large volumes of real-world data often exhibit inhomogeneities: vertically in the form of correlated or independent dimensions and horizontally in the form of clustered or scattered data items. In essence, these inhomogeneities form the patterns in the data that researchers are trying to find and understand. Sophisticated statistical methods are available to reveal these patterns, however, the visualization of their outcomes is mostly still performed in a one-view-fits-all manner, In contrast, our novel visualization approach, VisBricks, acknowledges the inhomogeneity of the data and the need for different visualizations that suit the individual characteristics of the different data subsets. The overall visualization of the entire data set is patched together from smaller visualizations, there is one VisBrick for each cluster in each group of interdependent dimensions. Whereas the total impression of all VisBricks together gives a comprehensive high-level overview of the different groups of data, each VisBrick independently shows the details of the group of data it represents, State-of-the-art brushing and visual linking between all VisBricks furthermore allows the comparison of the groupings and the distribution of data items among them. In this paper, we introduce the VisBricks visualization concept, discuss its design rationale and implementation, and demonstrate its usefulness by applying it to a use case from the field of biomedicine.
G. Li, R. Palmer, M. DeLisi, G. Gopalakrishnan, R.M. Kirby.
“Formal Specification of MPI 2.0: Case Study in Specifying a Practical Concurrent Programming API,” In Science of Computer Programming, Vol. 76, pp. 65--81. 2011.
DOI: 10.1016/j.scico.2010.03.007
We describe the first formal specification of a non-trivial subset of MPI, the dominant communication API in high performance computing. Engineering a formal specification for a non-trivial concurrency API requires the right combination of rigor, executability, and traceability, while also serving as a smooth elaboration of a pre-existing informal specification. It also requires the modularization of reusable specification components to keep the length of the specification in check. Long-lived APIs such as MPI are not usually 'textbook minimalistic' because they support a diverse array of applications, a diverse community of users, and have efficient implementations over decades of computing hardware. We choose the TLA+ notation to write our specifications, and describe how we organized the specification of around 200 of the 300 MPI 2.0 functions. We detail a handful of these functions in this paper, and assess our specification with respect to the aforementioned requirements. We close with a description of possible approaches that may help render the act of writing, understanding, and validating the specifications of concurrency APIs much more productive.
J. Li, J. Li, D. Xiu.
“An Efficient Surrogate-based Method for Computing Rare Failure Probability,” In Journal of Computational Physics, Vol. 230, No. 24, pp. 8683--8697. 2011.
DOI: 10.1016/j.jcp.2011.08.008
In this paper, we present an efficient numerical method for evaluating rare failure probability. The method is based on a recently developed surrogate-based method from Li and Xiu [J. Li, D. Xiu, Evaluation of failure probability via surrogate models, J. Comput. Phys. 229 (2010) 8966–8980] for failure probability computation. The method by Li and Xiu is of hybrid nature, in the sense that samples of both the surrogate model and the true physical model are used, and its efficiency gain relies on using only very few samples of the true model. Here we extend the capability of the method to rare probability computation by using the idea of importance sampling (IS). In particular, we employ cross-entropy (CE) method, which is an effective method to determine the biasing distribution in IS. We demonstrate that, by combining with the CE method, a surrogate-based IS algorithm can be constructed and is highly efficient for rare failure probability computation—it incurs much reduced simulation efforts compared to the traditional CE-IS method. In many cases, the new method is capable of capturing failure probability as small as 10-12 ~ 10-6 with only several hundreds samples.
Keywords: Rare events, Failure probability, Importance sampling, Cross-entropy
L. Lins, D. Koop, J. Freire, C.T. Silva.
“DEFOG: A System for Data-Backed Visual Composition,” SCI Technical Report, No. UUSCI-2011-003, SCI Institute, University of Utah, 2011.
Page 61 of 144
